Analyze primer properties
Biomedical Genomics Workbench can calculate and display the properties of predefined primers and probes:
Toolbox | Primers and Probes (![]() )|
Analyze Primer Properties (
)|
Analyze Primer Properties (![]() )
)
If a sequence was selected before choosing the Toolbox action, this sequence is now listed in the Selected Elements window of the dialog. Use the arrows to add or remove a sequence from the selected elements. (Primers are represented as DNA sequences in the Navigation Area).
Clicking Next generates the dialog seen in figure 34.16:
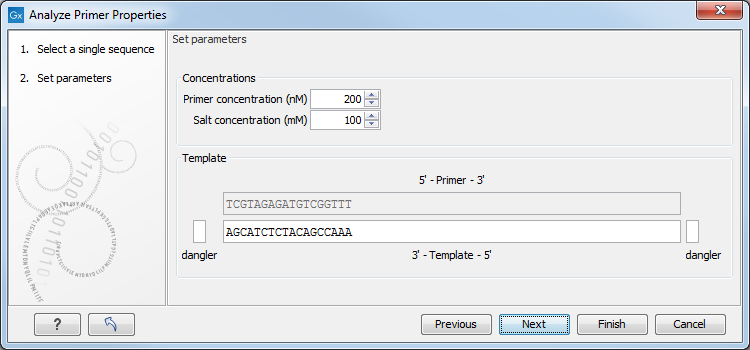
Figure 34.16: The parameters for analyzing primer properties.
In the Concentrations panel a number of parameters can be specified concerning the reaction mixture and which influence melting temperatures
- Primer concentration. Specifies the concentration of primers and probes in units of nanomoles (
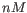 )
)
- Salt concentration. Specifies the concentration of monovalent cations (
![$ [NA^+]$](img247.gif) ,
, ![$ [K^+]$](img248.gif) and equivalents) in units of millimoles (
and equivalents) in units of millimoles (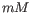 )
)
In the Template panel the sequences of the chosen primer and the template sequence are shown. The template sequence is as default set to the reverse complement of the primer sequence i.e. as perfectly base-pairing. However, it is possible to edit the template to introduce mismatches which may affect the melting temperature. At each side of the template sequence a text field is shown. Here, the dangling ends of the template sequence can be specified. These may have an important affect on the melting temperature [Bommarito et al., 2000]
Click Finish to start the tool. The result is shown in figure 34.17:
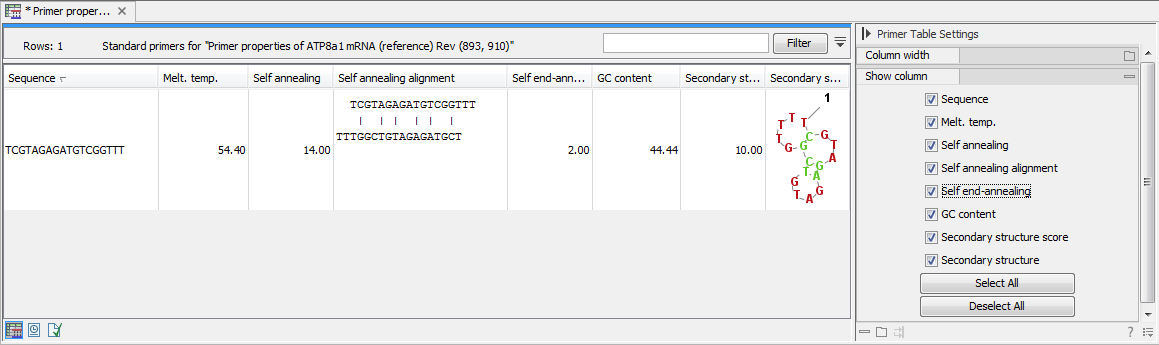
Figure 34.17: Properties of a primer.
In the Side Panel you can specify the information to display about the primer. The information parameters of the primer properties table are explained in the Standard PCR output table section.
