Differential Expression for RNA-Seq
The Differential Expression for RNA-Seq tool performs a statistical differential expression test for a set of Expression Tracks. It uses multi-factorial statistics based on a negative binomial GLM. The tool supports paired designs and can control for batch effects. The statistical analysis is described in more detail in The statistical model.
To run the Differential Expression for RNA-Seq analysis:
Toolbox | RNA-Seq Analysis | Differential Expression for RNA-Seq
Select a number of Expression tracks (![]() ) and click Next. For Expression Tracks (TE), the values used as input are "Total transcript reads".
For Gene Expression Tracks (GE), the values used depend on whether an eukaryotic or prokaryotic organism is analyzed, i.e. if the option "Genome annotated with Genes and transcripts" or "Genome annotated with Genes only" is used. For Eukaryotes the values are "Total Exon Reads", whereas for Prokaryotes the values are "Total Gene Reads".
) and click Next. For Expression Tracks (TE), the values used as input are "Total transcript reads".
For Gene Expression Tracks (GE), the values used depend on whether an eukaryotic or prokaryotic organism is analyzed, i.e. if the option "Genome annotated with Genes and transcripts" or "Genome annotated with Genes only" is used. For Eukaryotes the values are "Total Exon Reads", whereas for Prokaryotes the values are "Total Gene Reads".
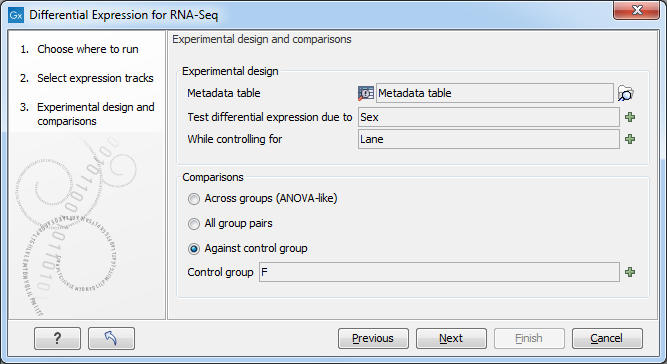
Figure 29.27: Setting up the experimental design and comparisons.
This will display the wizard shown in figure 29.27.
In the Experimental design panel, a Metadata table must be selected that describes the factors and groups for all the samples.
- Metadata table The metadata table describing the factors for the selected Expression tracks.
- Test differential expression due to Specify the one factor differential expression is tested for.
- While controlling for Specify confounding factors, i.e., factors that are not of primary interest, but may affect gene expression.
The Comparisons panel determines the number and type of statistical comparison tracks output by the tool (see Output of the Differential Expression for RNA-Seq tool for more details).
|
How many replicates do I need? The Differential Expression for RNA-Seq tool is capable of running without replicates, but this is not recommended and the results should be treated with caution. In general it is desirable to have as many biological replicates as possible - typically at least |
Subsections
- The statistical model
- Output of the Differential Expression for RNA-Seq tool
- Statistical comparison tracks
- The volcano plot
