Binding parameters
This opens the dialog displayed in figure 34.18:
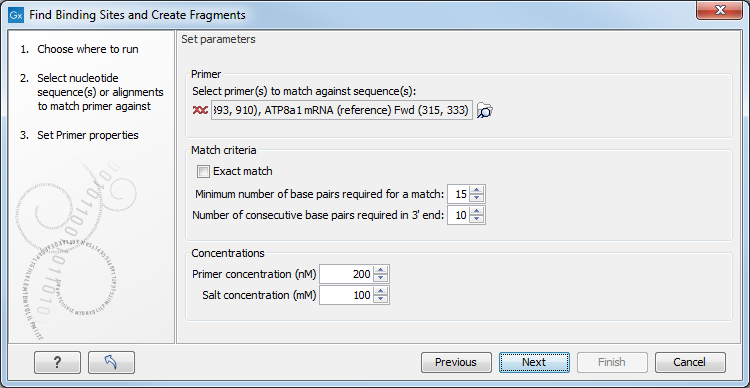
Figure 34.18: Search parameters for finding primer binding sites.
At the top, select one or more primers by clicking the browse (![]() ) button. In Biomedical Genomics Workbench, primers are just DNA sequences like any other, but there is a filter on the length of the sequence. Only sequences up to 400 bp can be added.
) button. In Biomedical Genomics Workbench, primers are just DNA sequences like any other, but there is a filter on the length of the sequence. Only sequences up to 400 bp can be added.
The Match criteria for matching a primer to a sequence are:
- Exact match. Choose only to consider exact matches of the primer, i.e. all positions must base pair with the template.
- Minimum number of base pairs required for a match. How many nucleotides of the primer that must base pair to the sequence in order to cause priming/mispriming.
- Number of consecutive base pairs required in 3' end. How many consecutive 3' end base pairs in the primer that MUST be present for priming/mispriming to occur. This option is included since 3' terminal base pairs are known to be essential for priming to occur.
Below the match settings, you can adjust Concentrations concerning the reaction mixture. This is used when reporting melting temperatures for the primers.
- Primer concentration. Specifies the concentration of primers and probes in units of nanomoles (
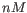 )
)
- Salt concentration. Specifies the concentration of monovalent cations (
![$ [NA^+]$](img247.gif) ,
, ![$ [K^+]$](img248.gif) and equivalents) in units of millimoles (
and equivalents) in units of millimoles (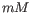 )
)
