Import RNA spike-in controls
The Biomedical Genomics Workbench has a dedicated tool for importing RNA spike-in control data:
Import | RNA Spike-ins
The wizard offers the option to import a standard ERCC file as provided by Thermo Fisher Scientific, or a custom made one (figure 6.19).
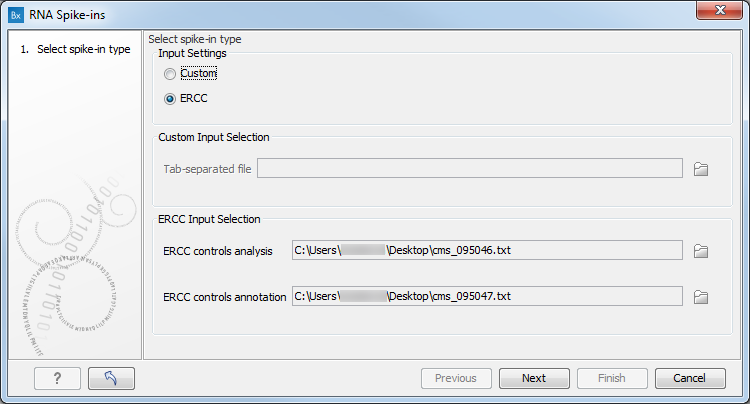
Figure 6.19: The RNA spike-in controls importer.
To import a standard ERCC file, look for ERCC Controls Analysis and ERCC Control Annotation files on the Thermo Fisher Scientific website, download both *.txt files on your computer, and start the importer. Select the option "ERCC" and specify the location of the analysis and annotation files in the relevant "ERCC Input Selection" fields at the bottom of the wizard.
For custom-made spike-in controls, choose the "Custom" option and specify in the "Custom Input Selection" field a tab-separated file (*.tsv or *.txt) containing the spike-in data organized as such: sequence name in the first column, nucleotide sequence in the second column, followed by as many columns as necessary to contain the concentrations of the spike-in measures in attomoles/microliters. Concentrations must not contain commas: write 15000 instead of 15,000. Remove any white space and save the table as a tab-separated TSV or TXT file on your computer.
It is also possible to import Lexogen Spike-in RNA Variant Control Mixes by modifying the SIRV files to fit the custom file requirements. Download the SIRV sequence design overview (XLSX) from the Lexogen website and open it in Excel. In the annotation column, "c" designate the data that should be imported ("i" is under-annotated while "0" is over-annotated). Filter the table to only keep the rows having a 1 in the "c" column, then keep only - and in that order - the sequence name, nucleotide sequence and concentration columns of the remaining rows. Reformat the values to numerical values in attomoles/microliters before saving the table as a *.tsv file. Import the file in the workbench using the "Custom" option.
Once a spike-in file is specified, click Next and choose to Save the file in the Navigation Area for later use in RNA-Seq Analysis.
