Genome browser
This chapter explains how to visualize tracks, how to retrieve reference data and finally how to perform generic comparisons between tracks.
The genome browser is the graphical interface where tracks can be presented alone or together with other tracks. Tracks are the fundamental building blocks for data analysis in the Biomedical Genomics Workbench and provide a unified framework for the visualization, comparison and analysis of genome-scale studies.
In tracks, all information is tied to genomic positions. A central coordinate-system is provided by a reference genome, which allows different types of data or results for different samples to be seen and analyzed together.
Different types of data are represented in different types of tracks, and each type of track has its own particular editors. An example of a paired-end mapping read-track displaying reads and coverage is shown in figure 19.1.
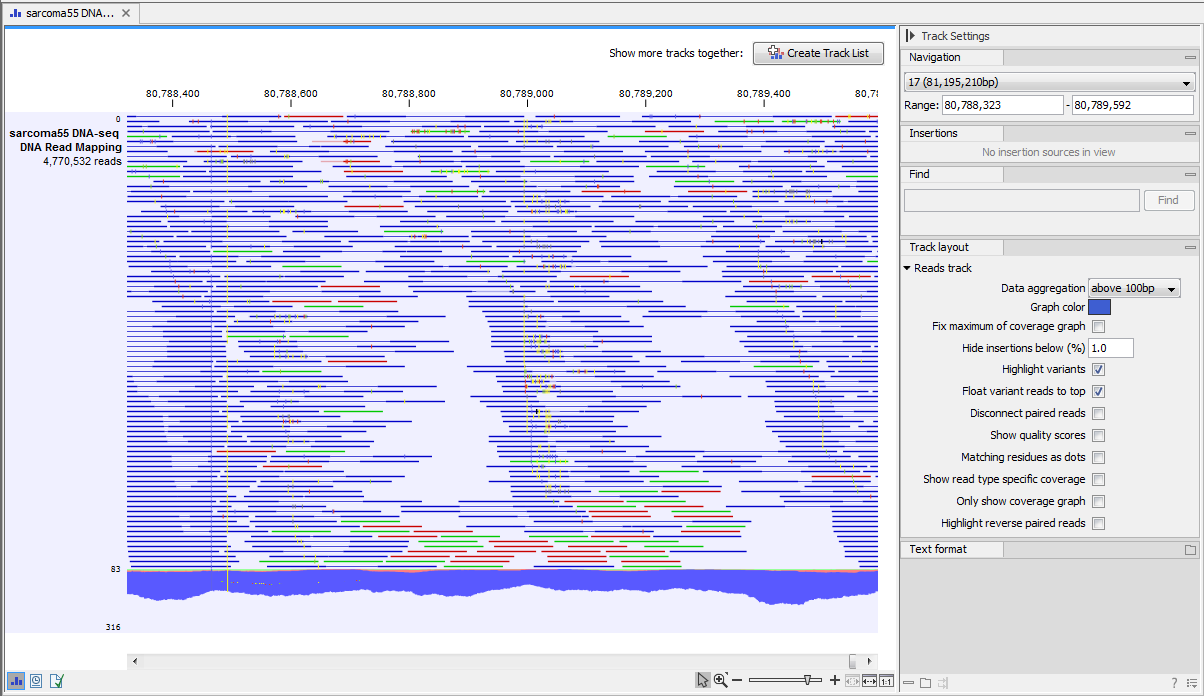
Figure 19.1: A paired-end mapping read-track opened, displaying reads and coverage. On the top right, the button for creating a Track List is visible. On the right is the Side Panel.
Subsections
