Dynamic restriction sites
If you open a sequence, a sequence list etc, you will find a Restriction Sites
section in the Side Panel.
Restriction sites can be shown on the sequence as colored triangles and lines (figure 32.1): check the "Show" option on top of the Restriction sites section, then specify the enzymes that should be displayed.
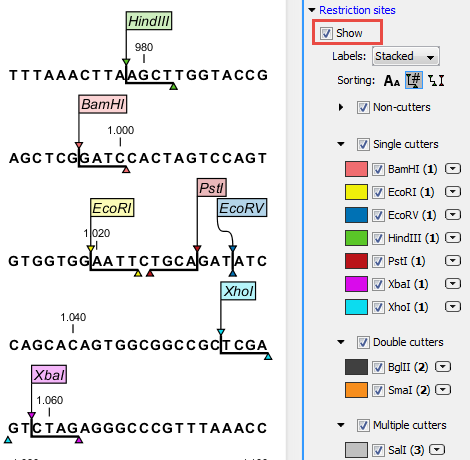
Figure 32.1: Showing restriction sites of ten restriction enzymes.
The color of the restriction enzyme can be changed by clicking the colored box next to the enzyme's name. The name of the enzyme can also be shown next to the restriction site by selecting Show name flags above the list of restriction enzymes.
There is also an option to specify how the Labels shown be shown:
- No labels. This will just display the cut site with no information about the name of the enzyme. Placing the mouse button on the cut site will reveal this information as a tool tip.
- Flag. This will place a flag just above the sequence with the enzyme name (see an example in figure 32.2). Note that this option will make it hard to see when several cut sites are located close to each other. In the circular view, this option is replaced by the Radial option.
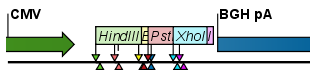
Figure 32.2: Restriction site labels shown as flags. - Radial. This option is only available in the circular view. It will place the restriction site labels as close to the cut site as possible (see an example in figure 32.3).
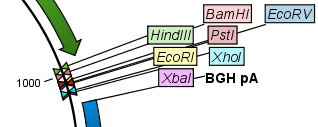
Figure 32.3: Restriction site labels in radial layout. - Stacked. This is similar to the flag option for linear sequence views, but it will stack the labels so that all enzymes are shown. For circular views, it will align all the labels on each side of the circle. This can be useful for clearly seeing the order of the cut sites when they are located closely together (see an example in figure 32.4).
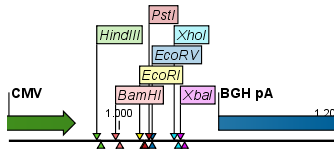
Figure 32.4: Restriction site labels stacked.
Note that in a circular view, the Stacked and Radial options also affect the layout of annotations.
Just above the list of enzymes, three buttons can be used for sorting the list (see figure 32.5).
![]()
Figure 32.5: Buttons to sort restriction enzymes.
- Sort enzymes alphabetically (
 ).
Clicking this button will sort the list of enzymes alphabetically.
).
Clicking this button will sort the list of enzymes alphabetically.
- Sort enzymes by number of restriction sites (
 ).
This will divide the enzymes into four groups:
).
This will divide the enzymes into four groups:
- Non-cutters.
- Single cutters.
- Double cutters.
- Multiple cutters.
- Sort enzymes by overhang (
 ).
This will divide the enzymes into three groups:
).
This will divide the enzymes into three groups:
- Blunt. Enzymes cutting both strands at the same position.
- 3'. Enzymes producing an overhang at the 3' end.
- 5'. Enzymes producing an overhang at the 5' end.
Subsections
