Genome model
The Genotype track genome model consists of four elements as shown in figure 10.1. A variant locus and an allele variant always have a conceptual component and may have a sample specific component. Haplotype alleles represent instances of allele variants in a sample, and when haplotype alleles are present in the same DNA molecule they form a haplotype together (i.e. the haplotype alleles are phased).
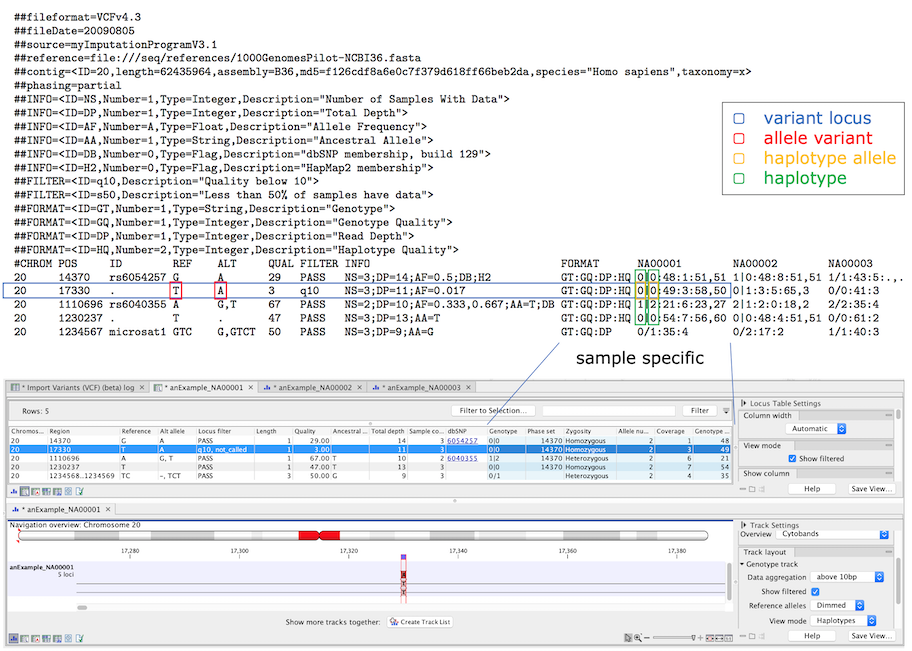
Figure 10.1: The four elements in the genome model and their relation to VCF. The variant locus has a number of allele variants that in a sample has a number of haplotype alleles, and phased haplotype alleles at several loci form a haplotype. In the lower part, the VCF has been imported and a Genotype track locus table and track view are showing a filtered locus. Sample specific annotations are colored blue in the Genotype track tables.
Filtering
Just as it is possible to specify filters in the FILTER column of a VCF file, it is possible to apply filters to a locus in the Genotype track. Additionally, allele variants and haplotype alleles can have filters directly applied.Elements can also have indirectly applicable filters if either the parent element or all child elements have applied filters. For example an allele variant has indirectly applicable filters if all constituent haplotype alleles have applied filters.
That a filter applies indicates that the element should be ignored and is not considered to be present in the sample. Elements with applicable filters are not shown by default, though can be displayed by enabling 'Show filtered' in the side panels of tables and track view (figure 10.1).
Read about variant filtering in the Microhaplotype Caller under Microhaplotype Caller - Filtering.
Complex representation
In a sample Genotype track, multiple overlapping loci together constitute a complex locus. For haplotype alleles in different overlapping loci that describe the same haplotype, there is typically a number of loci with the preferred representation of the allele, and the remaining loci have overlap alleles for that haplotype. Overlap alleles are marked with the 'Overlap allele' annotation (see allele table). The actual sequence may be unavailable for the overlap alleles, in which case they are represented as a chosen overlap variant. The overlap allele representation in the Genotype track is 'reference overlap', meaning that overlap alleles where the actual sequence is unavailable are represented as the reference allele variant. Other used overlap allele representations are star alleles and symbolic alleles, however these options are not available in the Genotype track yet.
