Use Genome as Result
The Use Genome as Result tool is part of the Map to Specified Reference workflow scenario and is not necessarily intended to be used as a single tool by users. Its function, at the last step of the Map to Specified Reference workflow, is double: it adds the name of the reference genome used for the re-mapping to the 'role' of the input files (for example the role "mapping report" will become "NZ_CP014971 mapping report", where NZ_CP014971 is the name of the reference used to re-map). It also creates an extra column in the Result Metadata Table whose header is the name of the common reference that was used for the re-mapping (here NZ_CP014971). This extra column makes it possible to distinguish between read mappings that were generated at different time points as well as in different runs of the workflow, despite using the same genome reference.
The tool can take multiple elements as input, and each will have its metadata role changed to include the name of reference sequence in addition to the original role value. Relevant elements can be selected individually, or you can select folders by right-clicking on the folder value and selecting Add folder contents (it will select all elements in that folder), or select folder recursively by right-clicking on the folder value and selecting Add folder contents (recursively). In this last case all elements of the folder, including elements contained in subfolders, will be selected (see figure 11.15).
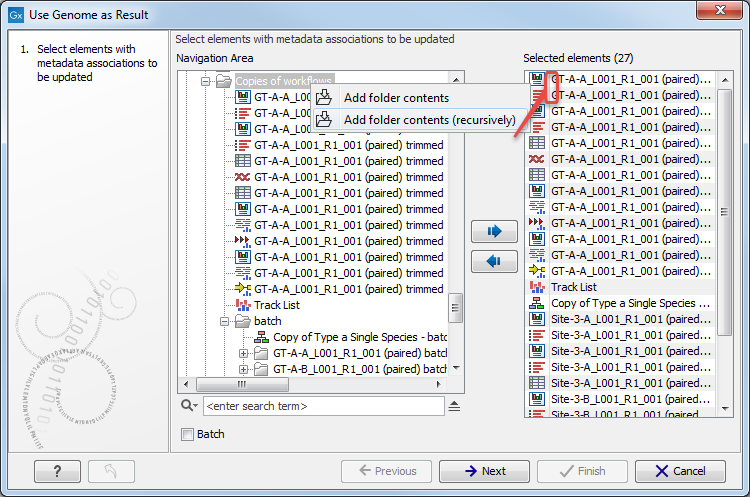
Figure 11.15: Select recursively elements with metadata associations to be updated. Here the selected elements are the 16 of the folder 'Copies of workflows' as well as the 9 from the subfolder called 'batch'.
In the second dialog, select the relevant read mapping, i.e., the read mapping that was created using the common reference you want to annotate the roles with (figure 11.16).
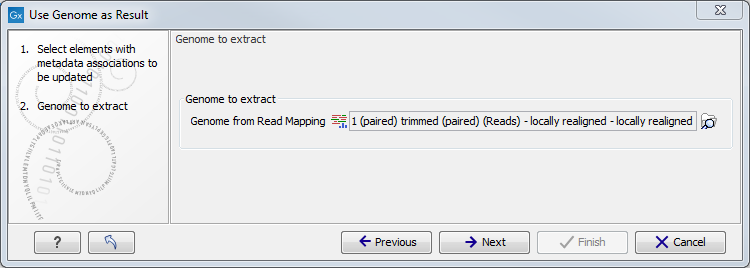
Figure 11.16: Specification of the read mapping to be associated with genome metadata.
In addition to changing the role name, the tool creates a new column named after the selected reference sequence in the Result Metadata Table. This column indicates whether data has been analysed using this reference or not (For an example see column "NZ_CP014971" in figure 11.17). To filter for all possible elements that were generated using this sequence as reference data, open the filter banner by clicking on the icon (![]() ) next to the Filter button. In the first drop down menu, choose the column whose header is the reference sequence of interest. In the second drop down menu, choose the term "contains", and in the third window, write "true". This will filter for all the elements with a tick in the reference sequence column, as can be seen on the figure 11.17.
) next to the Filter button. In the first drop down menu, choose the column whose header is the reference sequence of interest. In the second drop down menu, choose the term "contains", and in the third window, write "true". This will filter for all the elements with a tick in the reference sequence column, as can be seen on the figure 11.17.
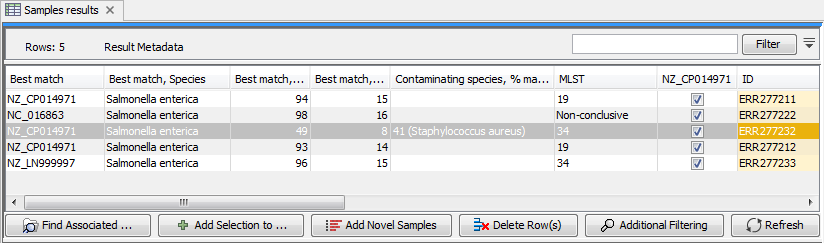
Figure 11.17: Filter for elements who share a tick in the column newly generated by the Use Genome as Result tool
