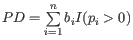Alpha diversity measures
The available diversity measures are:
- Total number: The number of features (e.g. OTUs when doing out clustering, GO terms when building functional profiles or organisms when performing taxonomic profiling) observed in the sample.
- Chao 1 bias-corrected:
 .
.
- Chao 1:
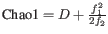 .
.
- Simpson's index:
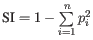 .
.
- Shannon entropy:
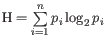 .
.
Note that Chao-based methods deal with singletons and doubletons, i.e., rows with exactly one or two reads (counts) associated. These measures are thus not available for whole metagenome taxonomic profiles that are characterized by coverage estimate.
The following distances are also available:
- Phylogenetic diversity: