Find Best Matches using K-mer Spectra
The Find Best Matches using K-mer Spectra tool is inspired by Hasman2013 and Larsen2014 and enables identification of the best matching reference among a specified reference sequence list. This section intends to describe the tool if you would like to use it as such. However, we recommend to use the Type a Known Species or Type among Multiple Species template workflows instead as described in the chapter Workflow templates.
To identify best matching bacterial genome reference, go to:
Typing and Epidemiology (beta) (![]() ) | Find Best Matches using K-mer Spectra (
) | Find Best Matches using K-mer Spectra (![]() )
)
Select the sequences you want want to find a best match sequence for (figure 13.1).
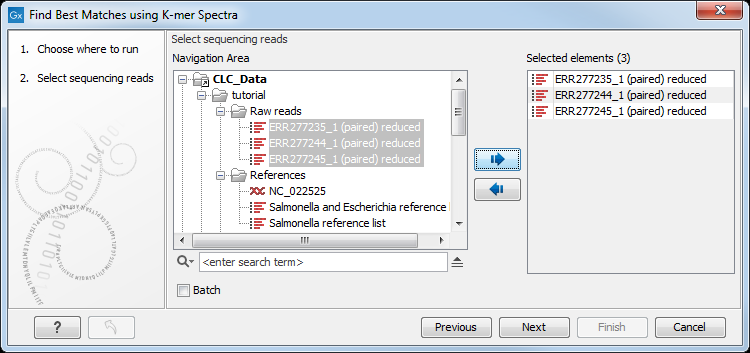
Figure 13.1: To identify best matching reference, specification of read file is the first step.
Select then a reference database, and specify the following settings (figure 13.2).
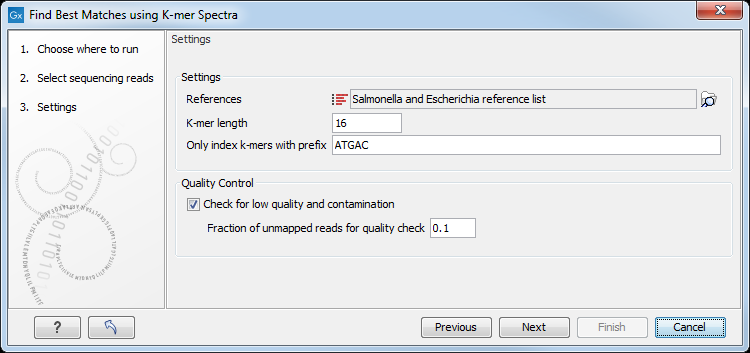
Figure 13.2: Specify reference list to search across.
- References may be a single- or multiple list(s) of sequences. It is for example possible to use the full NCBI's bacterial genomes database, or subset(s) of it.
- K-mer length is the fixed number (k) of DNA bases to search across.
- Only index k-mers with prefix allows specification of the initial bases of the k-mer sequence to limit the search space.
- Check for low quality and contamination will perform a quality check of the input data and identify potential contaminations.
- Fraction of unmapped reads for quality check defines the contamination tolerance as the fraction of the total number of reads not mapping to the best reference.
In the last wizard window, the tool provides the following output options (figure 13.3).
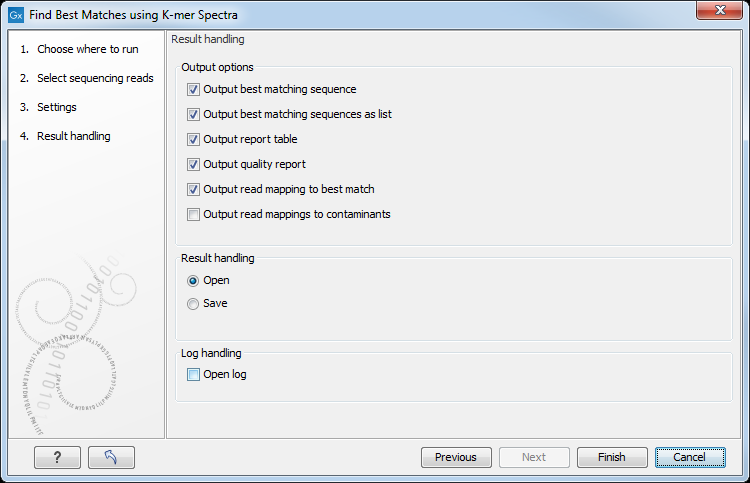
Figure 13.3: Choose your output option before saving your results.
- Output Best Matching Sequence is the best matching genome within the provided reference sequence list(s).
- Output Best Matching Sequences as a List includes the best matching genomes ordered with the best matching reference sequence first. The list is capped at 100 entries. Content is the same as in the Output Report Table.
- Output Report Table represents the best matching sequence. It lists all significantly matching references including various statistical values (as described in Hasman2013 and Larsen2014). The list is capped at 100 entries and the column headers are defined as such:
- Score Numbers of k-mers from the database seen in the reads.
- Expected The expected value, i.e., what score should been for the Z-score to be 0 and thus the P-value to be 1.
- Z Calculated Z-score.
- P Z-score translated to two-sided P-value.
- P, corrected P-value with Bonferroni correction.
- Output Quality Report gives a report with some statistics on possible contaminations and coverage reports for the read mappings. This option is available if the option Check for low quality and contamination was selected in the first wizard window. This report contains the metadata:
- Best match, % mapped Percent of reads mapping to the best matching reference.
- Contaminating species, % mapped (taxonomy info) Percent of mapping reads and the most specific accessible taxonomy information for the most probable contaminant.
- Output read mapping to best match gives the mapping of the reads to the best matching reference. This option is available if the option Check for low quality and contamination was selected in the first wizard window.
- Output read mapping to contaminants if a contamination is detected, this generates the mapping of the reads (which do not map to the best reference) to the probable contaminants. This option is available if the option Check for low quality and contamination was selected in the first wizard window.
In cases where the tool stops with a warning that good references were not found, you should download a new set of references for the organisms of interest and re-run the workflow.
To add the obtained best match to a Result Metadata Table, see the section Add to Result Metadata Table. Note that Best match results are added automatically to Result Metadata Table when using the template Type a Known Species and/or Type among Multiple Species workflow(s) or their customized versions.
Note that in rare instances, the lists of references found in the Output Best Matching Sequences as a List and Output Quality Report may differ. The reason is that the former list is compiled based on a "Winner takes all" based count of K-mers which attributes all uniquely found K-mers only to the reference with the highest Z-score,. The latter list however is produced by removing all reads mapping to the best matching reference and using the remaining reads as a basis for determining the next best match. Thus, in the second round the pool of K-mers has been altered, and some K-mers that determined the Z-score of the original second-best match may have been removed.
Once results from the Find Best Matches using K-mer Spectra tool are added to the Result Metadata Table, extra columns are present in the table, including the taxonomy of the best matching references. In addition, in case the quality control was activated, the table will include the percentage of reads mapping to the best reference and the most probable contaminating species (see figure 13.4).

Figure 13.4: Taxonomy of the best matching reference and quality
information is shown in the Metadata Result Table.
