Find Resistance with ShortBRED
This tool allows you to detect and quantify the presence of antibiotic resistance (AR) genes by running DIAMOND with a database consisting of peptide marker sequences which represent the genes of interest. This marker database can be downloaded using the tool Download Resistance Database. The Find Resistance with ShortBRED tool works similarly to the quantify step of ShortBRED, a public bioinformatics pipeline and resource. To learn more about the ShortBRED-Quantify tool [Kaminski et al., 2015], see http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004557.
Find Resistance with ShortBRED quantifies the presence of Antibiotic Resistance (AR) genes in a sample of NGS short reads. It is possible to output a sequence list containing all the input reads which contained a marker (each read in the output is annotated with metadata describing the phenotype detected in the read).
To start the tool, go to: Metagenomics | Drug Resistance Analysis | Find Resistance with ShortBRED.
The tool accepts a nucleotide sequence or sequence list as input.
In the next dialog (figure 8.6), several parameters are available:
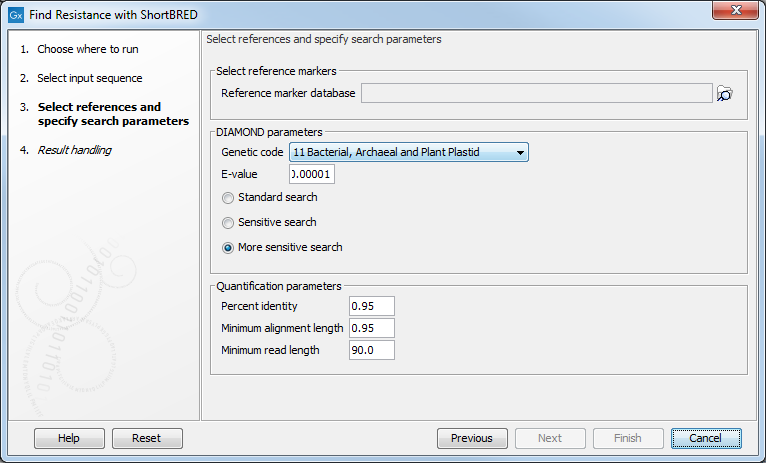
Figure 8.6: References and search parameters.
- Reference sequence database: select the AR marker database to use (downloaded with the tool Download Resistance Database).
- Genetic code: select the genetic code to use when DIAMOND translates the nucleotide sequences to proteins.
- E-value: specify an expectation value to use as threshold for qualifying hits with DIAMOND.
- Standard search: executes DIAMOND in its default mode.
- Sensitive search: executes DIAMOND in its sensitive mode.
- More sensitive search: executes DIAMOND in its most sensitive mode.
- Percent identity: defines a minimum threshold for the percent identity of an alignment. This value is used by Find Resistance with ShortBRED to determine whether a hit found by DIAMOND is sufficiently good to be validated as a true hit (equivalent to the parameter "-id" of the ShortBRED-Quantify tool).
- Minimum alignment length: the minimum length of the DIAMOND alignment. This value is used by Find Resistance with ShortBRED to determine whether a hit found by DIAMOND is sufficiently good to be validated as a true hit (equivalent to the parameter "-pctlength" of the ShortBRED-Quantify tool - see citation at the end of this section).
- Minimum read length: the minimum read length. This value is used by Find Resistance with ShortBRED to determine whether a read is long enough to be processed by Find Resistance with ShortBRED (equivalent to the parameter "-minreadBP" of the ShortBRED-Quantify tool).
The Find Resistance with ShortBRED tool will output an abundance table, an optional report, and an optional sequence list: the abundance table summarizes the abundance of each marker, i.e., it reports the number of times a given marker is found in the input reads. The abundance is also reported in units of RPKM, referred to as the normalized abundance, which are calculated in the same way as is done by ShortBRED-Quantify. It is possible to aggregate the abundance by gene name and resistance phenotype to get the abundance at each of these levels.
The optional report output summarizes the number of detected AR genes, the number of reads and the number of unique reads for each resistance phenotype. It also contains some general information about the input sample and the AR marker database.
The optional sequence list output contains all reads from the input sample which were found to contain one of the AR marker sequences. Each read in the sequence list is annotated with metadata describing the phenotype detected in the read.
