Annotate Variants (WTS)
Using a variant track, annotation track, expression track or statistical comparison track, the Annotate Variants (WTS) ready-to-use workflow adds the following annotations:
- Gene names Adds names of genes whenever a variant is found within a known gene.
- mRNA Adds names of mRNA whenever a variant is found within a known transcript.
- CDS Adds names of CDS whenever a variant is found within a coding sequence.
- Amino acid changes Adds information about amino acid changes caused by the variants.
- Information from ClinVar Adds information about the relationships between human variations and their clinical significance.
- Information from dbSNP Common Adds information from the "Single Nucleotide Polymorphism Database", which is a general catalog of genome variation, including SNPs, multinucleotide polymorphisms (MNPs), insertions and deletions (indels), and short tandem repeats (STRs).
- PhastCons Conservation scores The conservation scores, in this case generated from a multiple alignment with a number of vertebrates, describe the level of nucleotide conservation in the region around each variant.
- Go to the toolbox and select the Annotate Variants (WTS) workflow. In the first wizard step, select the variant track, annotation track, expression track or statistical comparison track to annotate (figure 23.2).
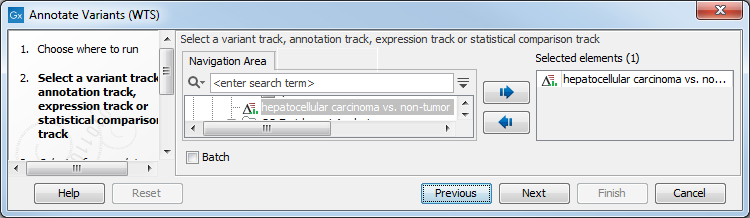
Figure 23.2: Select the variant, annotation, expression or statistical comparison track to annotate. - In the next dialog, you have to select which data set should be used to annotate variants (figure 23.3).
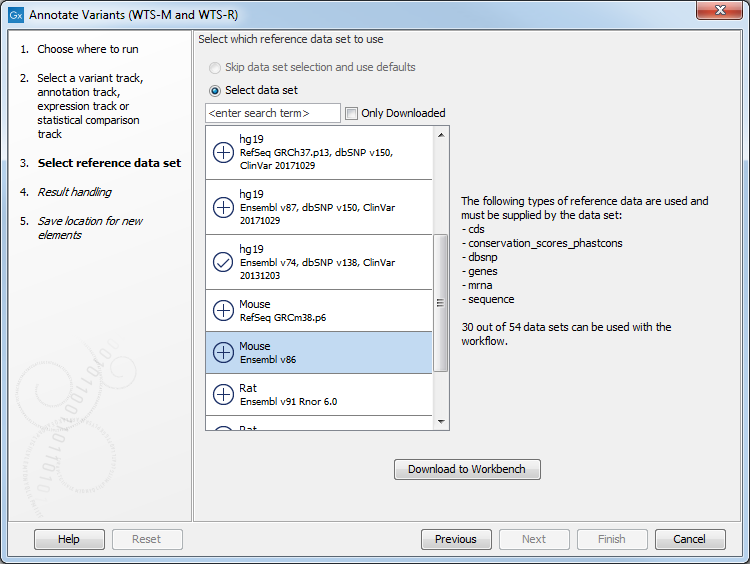
Figure 23.3: Choose the relevant reference Data Set to annotate. - If you are using the workflow for Human, you should specify which 1000 Genomes population to use (figure 23.4).
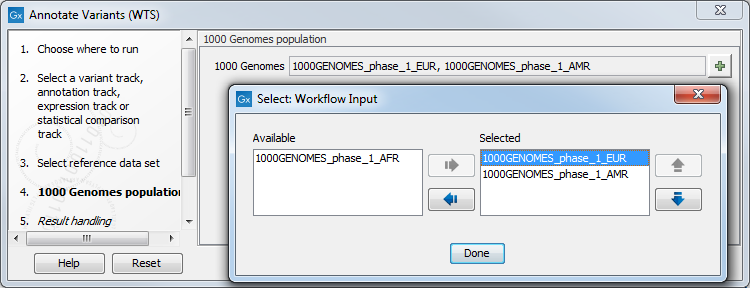
Figure 23.4: Select the relevant 1000 Genomes population(s). - In the last wizard step you can check the selected settings by clicking on the button labeled Preview All Parameters.
In the Preview All Parameters wizard you can only check the settings, and if you wish to make changes you have to use the Previous button from the wizard to edit parameters in the relevant windows.
- Choose to Save your results and click on the button labeled Finish.
The following outputs are generated:
- Annotated Variants (
 ) Annotation track showing the variants. Hold the mouse over one of the variants or right-clicking on the variant. A tooltip will appear with detailed information about the variant.
) Annotation track showing the variants. Hold the mouse over one of the variants or right-clicking on the variant. A tooltip will appear with detailed information about the variant.
- Track List Annotated Variants (
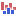 ) A collection of tracks presented together. Shows the annotated variant track together with the human reference sequence, genes, transcripts, coding regions, and variants detected in dbSNP Common, ClinVar, 1000 Genomes, and PhastCons conservation scores (see figure 23.5).
) A collection of tracks presented together. Shows the annotated variant track together with the human reference sequence, genes, transcripts, coding regions, and variants detected in dbSNP Common, ClinVar, 1000 Genomes, and PhastCons conservation scores (see figure 23.5).
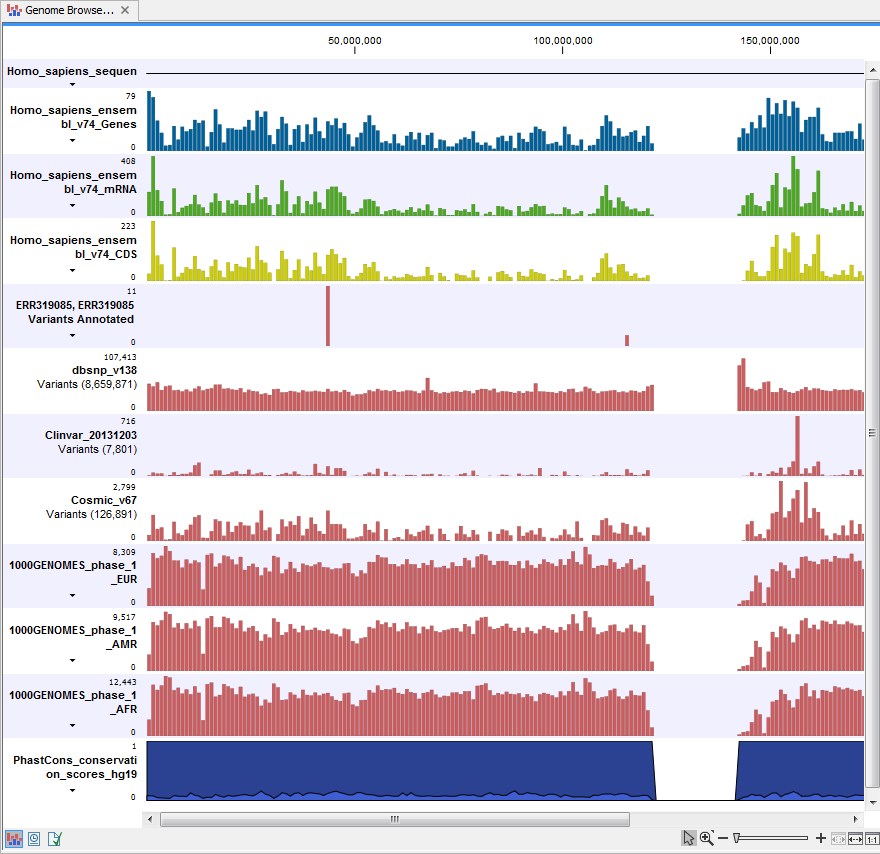
Figure 23.5: The output from the Annotate Variants (WTS) ready-to-use workflow is a track list containing individual tracks for all added annotations.
Note! Please be aware that if you delete the annotated variant track, this track will also disappear from the track list. On the other hand, it is possible to add tracks to the Track List by dragging the track directly from the Navigation Area to the Track List view.
Open the annotated variant track as a table; it includes all variants and the added information/annotations (see figure 23.6). The table and the Track List are linked; if you click on an entry in the table, this particular position in the genome will automatically be brought into focus in the Track List view.
Figure 23.6: The variant track in the Track List is open as a table in split view.
You may be met with a warning as shown in figure 23.7. This is simply a warning telling you that it may take some time to create the table if you are working with tracks containing large amounts of annotations. Please note that in case none of the variants are present in ClinVar or dbSNP Common, the corresponding annotation column headers are missing from the result.
Figure 23.7: Warning that appears when you work with tracks containing many annotations.
Adding information from other sources may help you identify interesting candidate variants for further research. For example, known common genetic variants (present in the HapMap database) or variants known to play a role in drug response or other relevant phenotypes (present in the ClinVar database) can easily be identified. Further, variants not found in the ClinVar database can be prioritized based on amino acid changes in case the variant causes changes on the amino acid level.
A high conservation level between different vertebrates or mammals, in the region containing the variant, can also be used to give a hint about whether a given variant is found in a region with an important functional role. If you would like to use the conservation scores to identify interesting variants, we recommend that variants with a conservation score of more than 0.9 (PhastCons score) is prioritized over variants with lower conservation scores.
It is possible to filter variants based on their annotations. This type of filtering can be facilitated using the table filter found at the top part of the table. If you are performing multiple experiments where you would like to use the exact same filter criteria, you can create a filter that can be saved and reused. To do this, use the Filter on Custom Criteria tool.
