Identify QIAseq DNA Somatic Variants with FLT3 Identification ready-to-use workflow
The Identify QIAseq DNA Somatic Variants with FLT3 Identification (Illumina) ready-to-use workflow supports the detection of FLT3 complex variants in addition to the somatic variant detection performed by the Identify QIAseq Somatic Variants (Illumina) workflows (see The Identify QIAseq DNA Variants ready-to-use workflows). Please note that parameters in the InDels and Structural Variants tools in the Identify QIAseq DNA Somatic Variants with FLT3 Identification (Illumina) workflow have been optimized to detect FLT3 internal tandem duplications, which may impact the ability to detect other variants.
This workflow is executed the same way as the Identify QIAseq DNA Variant workflows, and generates the same outputs, but with the addition of a variant track containing any detected FLT3 structural variant.
FLT3 is a gene situated at position 28577411-2874729 on the minus strand of chromosome 13 (hg19). This gene is known to include complex variability in terms of internal tandem duplications (ITDs) in the range of around 20 to a few hundred base pairs. In particular, the larger ITDs can be tricky to detect as read mappers can have problems mapping reads correctly, and downstream tools consequently are challenged in identifying the variant. To detect ITD variants within the FLT3 gene region, a second round of InDel detection, using the InDels and Structural Variant analysis tool, is included in this workflow. Count, coverage and frequency annotations are then added to the generated results using the Annotate Structural Variants tool (see Annotate Structural Variants).
The main purpose of the second instance of the InDels and Structural Variants algorithm is to pick up InDels and structural variants that the first instance of the local realignment did not manage to align into the reads. In addition, the first instance of the local realignment will have changed the read mapping, and the second instance of the InDels and Structural Variants algorithm hence may find InDels and structural variants that differ (most likely slightly) from those found by the first instance of the InDels and Structural Variants algorithm. The purpose of the second instance of the local realignment is to realign the reads in accordance with the InDels discovered in the second instance of the InDels and Structural Variants algorithm.
The detected FLT3 structural variants are reported in a variant track called Structural_variants_in_FLT3, which is saved to the Reports and Data folder. These variants can be exported to VCF format file, along with the other variants detected by the workflow.
The Identify QIAseq DNA Somatic Variants with FLT3 Identification ready-to-use workflow can be found at:
Ready-to-Use Workflows | QIAseq Panel Analysis (![]() ) | QIAseq Analysis workflows (
) | QIAseq Analysis workflows (![]() ) | Identify QIAseq DNA Somatic Variants with FLT3 Identification (Illumina)
) | Identify QIAseq DNA Somatic Variants with FLT3 Identification (Illumina)
This workflow can also be launched from the Analyze QIAseq Panels guide, described in The Analyze QIAseq Panels guide. It is available under the Targeted DNA tab for the Myeloid Neoplasms Panel (DHS-003Z) as well as the Comprehensive Cancer Panel (DHS-3501Z).
Most of the outputs generated by the Identify QIAseq DNA Somatic Variants with FLT3 Identification (Illumina) are described in Output from the Identify QIAseq DNA Variants workflows. One output is unique to the FLT3 workflow, a variant track with the prefix Structural_variants_in_FLT3. The Structural_variants_in_FLT3 track is included in the Track List, which allows visual inspection of detected FLT3 structural variants together with the mapped reads.
Please note that when inspecting the read mapping you may sometimes not be able to see the reads supporting a tandem duplication aligned nicely, but will only see evidence for the detected ITD in terms of reads with unaligned ends whose consensus sequence map back to the reference in a pattern consistent with the presence of an ITD. The unaligned ends are used by the Identify QIAseq DNA Somatic Variants with FLT3 Identification (Illumina) workflow to attempt to infer tandem duplications indirectly.
Due to the nature of FLT3 tandem duplications, the local realignment is not always able to recognize the inserted sequence and align the reads containing it properly. Whether or not the local realignment is successful depends on the particular composition of the reference sequence and the nature of alternate alleles in the vicinity of the tandem duplication. Figure 6.6 shows an example of a FLT3 internal tandem duplication that have been inferred from unaligned ends.
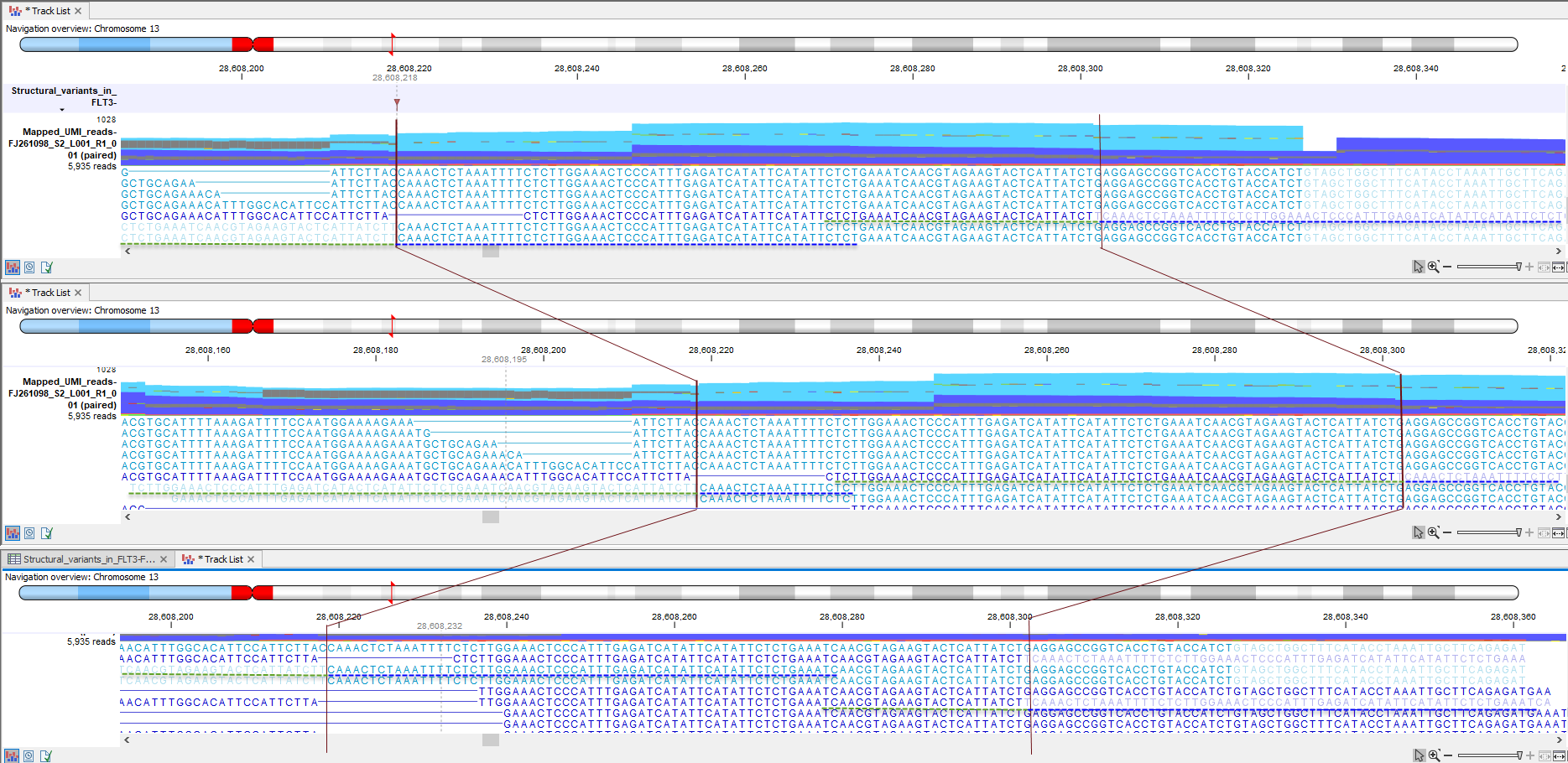
Figure 6.6: In this example the FLT3 tandem duplication was inferred indirectly from the unaligned ends and when looking at the read mapping the FLT3 is only seen as unaligned ends. The FLT3 tandem duplication is in a case like this not reported in the variant track, but is listed in the Structural Variants in FLT3 track. The three shown Track Lists are exactly the same but focusing on slightly different regions. In the top Track List the region with the tandem duplication is brought into focus (flanked by two horizontal red lines). In the Track List found in the middle, focus is on the left-most unaligned end, and at the bottom a Track List focusing on the right-most unaligned end is shown. Green dotted lines in each Track List highlight where the left-most unaligned end has an identical sequence in the aligned part of the read. Likewise, blue dotted lines in each Track List highlight where the right-most unaligned end has an identical sequence in the aligned part of the read
