RNA-Seq and Differential Gene Expression Analysis workflow
The RNA-Seq and Differential Gene Expression Analysis workflow includes the steps necessary to carry out RNA-Seq analyses on trimmed reads, followed by a differential expression analysis of the RNA-Seq data. This workflow also generates reports and visualizations.
The workflow calculates expression profile for each sample individually and then conducts a differential expression analysis of all the samples, grouped based on information provided when launching the workflow. The ability to run parts of the workflow on a per-sample basis, and other parts based on groups of samples, is possible due to the inclusion of Iterate and Collect and Distribute elements in the workflow design (Workflow control flow elements).
If possible, samples with known results should be used to test and optimize the workflow settings to fit your specific application.
Some common adjustments of this workflow are provided at the end of this section.
Requirements for this workflow
To run this workflow, you will need:
- Trimmed reads Reads can be trimmed using the Trim Reads tool (NGS Trim Reads tool) or the Prepare Raw Data template workflow (Prepare Raw Data).
- An Excel or CSV format file containing information about the samples to be analyzed A column containing a unique identifier of each sample is needed, as well as information on the factors relevant to the differential expression analysis must be provided in this file (e.g. treatment level, sex, etc.). See figure 12.70. The unique identifiers in this file must at least partially match the names of the input sequence elements so that information from this file can be linked with the relevant read data.
In this section, we assume sequence lists containing your trimmed reads are in a single folder.
Launching the workflow
The RNA-Seq and Differential Gene Expression Analysis workflow is at:
Toolbox | Template Workflows | Basic Workflow Designs (![]() )
| RNA-Seq and Differential Gene Expression Analysis (
)
| RNA-Seq and Differential Gene Expression Analysis (![]() )
)
Launch the workflow and step through the wizard.
- Select the sequence lists containing your trimmed reads and click on Next.
- Select your reference data set or select "Use the default reference data" to configure the reference data elements individually in subsequent wizard steps.
- Choose the "Use metadata" for defining the batch units, and then select the Excel or CSV format file containing information about your samples. The metadata column to specify for defining the batch units is one with a unique entry per sample, where that information at least partially match the names of the input sequence elements. This would commonly be an ID for the samples (figure 12.71).
- In the next step, you can review the batch units resulting from your selections above.
- If you did not select a reference data set in the earlier step, then in the following steps, you will be prompted to specify the reference data elements to use.
- The differential expression settings are then specified (figure 12.72).
- Finally select a location to save results to and press Finish.
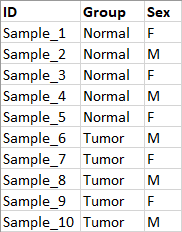
Figure 12.70: Metadata describing 10 samples from a tumor normal comparison experiment.
Tools in the workflow and outputs generated
The tools and outputs provided by this workflow are:
- QC for Sequencing Reads outputs a report that is useful for validating the quality of the reads after trimming.
- RNA-Seq Analysis outputs the Gene Expression Track and a Mapping Report per sample.
- Differential Expression for RNA-Seq produces Statistical Comparison Tracks. As default, the tool is set up to expect Whole Transcriptome RNA-Seq data and to compare all groups specified in the metadata. This can easily be adjusted in the relevant wizard steps when running the workflow or the configurations can be changed in a copy of the workflow. The experimental design will depend on the metadata that is provided.
- Create Venn Diagram for RNA-Seq outputs a Venn diagram for up to 5 groups.
- Gene Set Test requires a GOA database and outputs a pathway analysis. When not available simply remove this element from the workflow.
- Create Expression Browser collects and combines the Gene Expression Track and Statistical Comparison Tracks into a single table.
- PCA for RNA-Seq produces a plot of all the gene expression samples.
- Create Heat Map for RNA-Seq produces a heat map of the top 25 features in the samples.
- Track List outputs a Genome Browser view of the sequence, genes, mRNA, CDS, and the differential gene expression results in the form of Statistical Comparison Tracks.
The RNA-Seq expression analysis is conducted at gene level and the differential expressions is hence reported at gene level. The workflow can easily be modified to conduct transcript level expression analysis. To modify the workflow you need to Open Copy of Workflow (right-click the workflow and select this option).
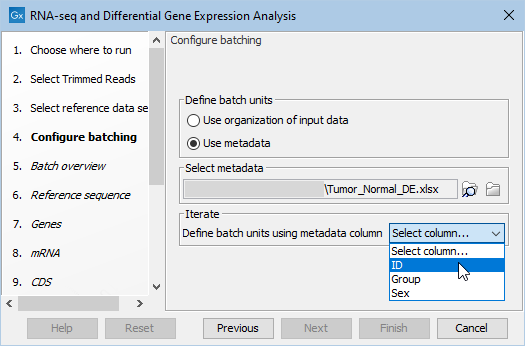
Figure 12.71: Atfer selection of the metadata file, select the samples identifyer in the drop-down menu.
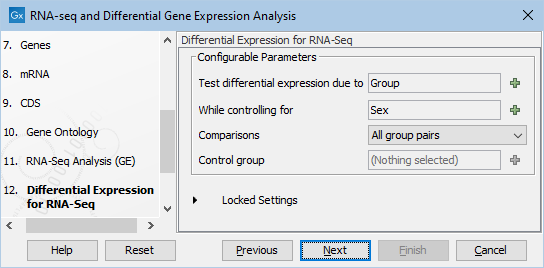
Figure 12.72: Specify based on the metadata how the differentital expression analysis should be conducted. In this case we chose to test differential expression due to Group (Tumor/Normal) while controling for Sex (F/M).
Workflow outputs resulting from analyses of all samples shown in figure 12.70 such as the PCA plot and the Venn Diagram are saved directly at the top level of the results folder. Outputs that are sample specific are organized in relevant sub folders, except expression tracks, see figure 12.73. Expression tracks for all samples are stored in the folder Gene Expression Tracks.
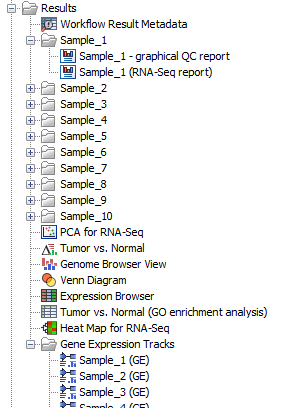
Figure 12.73: Overview of the outputs produced and how the folders are structured.
Customizing the RNA-Seq and Differential Gene Expression Analysis workflow design
Template workflows can be easily edited to add or remove analysis steps, change parameter settings, and so on. See Template workflows for information about how to open a template workflow for editing.
- Gene Set Test The workflow includes the Gene Set Test tools (REF), which requires a GOA database. If you do not have this for your species, remove the Gene Set Test element from the workflow.
- CDS track A CDS track is included as an input to the Create Track List element in the workflow. If you do not have CDS track for your reference genome, remove the CDS element from the workflow.
- Genes track and mRNA track The workflow is configured to make use of both these in the RNA-Seq Analysis step. They are also inputs to the Create Track List element. The RNA-Seq Analysis element can be configured with different "Reference types" and any reference tracks no longer needed can be removed from the workflow.
