Filter on Custom Criteria
The Filter on Custom Criteria tool can be used to identify and extract variants, annotations, or features that fulfill certain criteria. It will create a filtered copy of the input containing only the items that were not filtered away.
To run the Filter on Custom Criteria tool, go to:
Toolbox | Utility Tools (![]() ) | Filter on Custom Criteria (
) | Filter on Custom Criteria (![]() )
)
In the first step, select an input to be filtered. This can be a variant track (![]() ), annotation track (
), annotation track (![]() ), expression track (
), expression track (![]() ), statistical comparison track (
), statistical comparison track (![]() ), statistical comparison table (
), statistical comparison table (![]() ), miRNA seeds table (
), miRNA seeds table (![]() ), IsomiR table (
), IsomiR table (![]() ) among others. Click Next.
) among others. Click Next.
In the Filter criteria dialog, use the browse icon (![]() ) to select an element that contains at least all the criteria on which you want to filter the input. Once specified, click the Load Annotations button to create the drop down menu available in the filter field below. The drop down menu is constructed from the table view of the selected element.
) to select an element that contains at least all the criteria on which you want to filter the input. Once specified, click the Load Annotations button to create the drop down menu available in the filter field below. The drop down menu is constructed from the table view of the selected element.
An example of filter criteria is shown in figure 35.7. In this example, all variants that are not found on chromosome 1 and that are not homozygous are filtered away. As a result only variants that are homozygous and found on chromosome 1 are kept. The bottom filter field on the figure shows the various other filter criteria available after pressing the Load Annotations button.
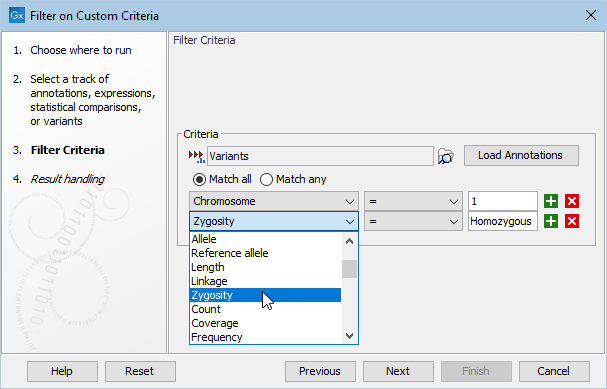
Figure 35.7: Create filter criteria to extract the variants of interest. In this example, the tool is configured to extract the homozygous variants found on chromosome 1.
The output from the Filter on Custom Criteria tool is a copy of the input containing only the items that satisfy the specified filter criteria.
