Detect and Refine Fusion Genes
Detect and Refine Fusion Genes finds fusion genes in a two-step process. The detect step identifies potential fusions and the refine step accumulates and evaluates the evidence for each fusion. Briefly, the detect step works by re-mapping the unaligned ends of reads and determining if these are consistent with a fusion. Fusions are identified from reads that must have an unaligned end close to an exon boundary that can be remapped close to another exon boundary. If the option for Detect fusions with novel exon boundaries has been enabled, the tool also considers reads that are far from an exon boundary and/or whose unaligned ends can be mapped far from an exon boundary in a second pass.
The refine step takes the fusions identified in the detect step, and re-counts the number of fusion crossing reads as well as the wildtype supporting reads using an RNA-Seq mapping against the wild type and fusion references. The fusion reference is an artificial reference sequence that "assumes" the detected fusions by generating new chromosomes corresponding to each fusion in addition to the original chromosomes (figure 31.51).
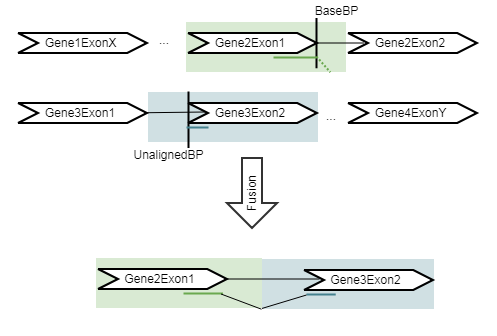
Figure 31.54: An artificial chromosome is created consisting of the vicinity of both ends of the fusion.
All reads are remapped to the artificial reference, with the expectation that reads that were used to detect the fusion will now map to the fusion transcript with a spliced read. In addition, some reads that did not originally map at all will now map to the artifical reference sequence, increasing evidence for the fusion event. The tool then calculates the Z-score and p-value using a binomial test.
The Detect and Refine Fusion Genes tool can be found in the Toolbox at:
Toolbox | RNA-Seq and Small RNA Analysis (![]() )| RNA-Seq Tools (
)| RNA-Seq Tools (![]() ) | Detect and Refine Fusion Genes (
) | Detect and Refine Fusion Genes (![]() )
)
The Detect and Refine Fusion Genes tool takes takes a sequence list (![]() ) as input (figure 31.52).
) as input (figure 31.52).
Figure 31.55: Select sequences.
In the next dialog (figure 31.53), specify the RNA-Seq reads track, as well as reference sequence, gene and mRNA tracks from the CLC_References folder of the Navigation Area. It is possible, but optional, to add a CDS or primer track to run the analysis.
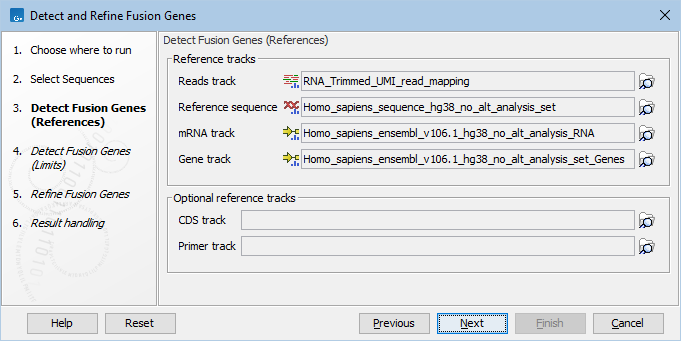
Figure 31.56: Specify reads track and references.
In the next dialog (figure 31.54), configure parameters for detecting fusion genes:
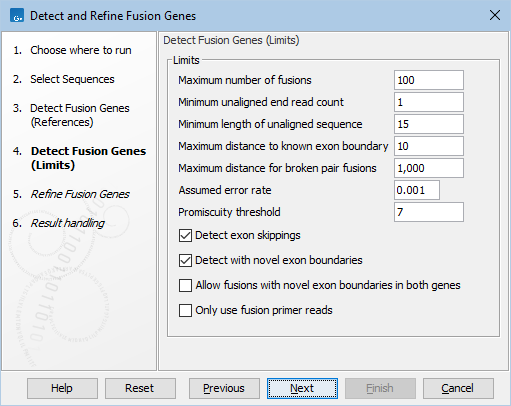
Figure 31.57: Default parameters for detecting fusion genes.
- Maximum number of fusions: The maximum number of putative fusions that will be evaluated. Multiple different possible fusion breakpoints between the same two genes count as 1 fusion.
- Minimum unaligned end read count: The minimum number of unaligned ends that must support a fusion, if lower than this number the fusion will not be considered in the refine step.
- Minimum length of unaligned sequence: Only unaligned ends longer than this will be used for detecting fusions.
- Maximum distance to known exon boundary: Reads with unaligned ends must map within this distance of a known exon boundary, and unaligned ends must map within this distance of another known exon boundary, to be recorded as supporting a fusion event.
Increasing this parameter counts reads that are further from a known exon boundary as if they fused at the boundary, which increases the signal for the fusion. However, increasing the parameter also decreases the resolution at which a fusion can be detected: for example, if "maximum distance to known exon boundary = 10" then two transcripts with exon boundaries 9nt apart will not be distinguished, and the tool will only produce artificial fusion transcripts for one of them, which can reduce the number of mapping reads in the refine step.
- Maximum distance for broken pairs fusions: The algorithm uses broken pairs to find additional support for fusion events. If a pair of reads originally mapped as a broken pair, but would not be considered broken if mapped across the fusion breakpoints (because the two reads in the pair then get close enough to each other), then that pair of reads supports the fusion event as "fusion spanning reads". The "Maximum distance for broken pairs fusions" parameter specifies how close to each other two broken pairs must map across the fusion breakpoints in order for them to be considered fusion spanning reads. This is usually set to the maximum paired end distance used for the Illumina import of reads.
- Assumed error rate: Value used to calculate Z-score and p-value.
- Promiscuity threshold: Only up to this number of fusion partners will be reported for a given gene.
This parameter does not limit the number of fusion breakpoints that can be reported between two genes, which is capped at 20 pairs of breakpoints:
We limit the number of breakpoint pairs between the same two genes by selecting the highest possible p-value threshold that admits at most 20 breakpoint pairs.
- Detect exon skippings: When enabled, same-gene fusions are reported.
- Detect with novel exon boundaries: When enabled, fusions beyond the distance set for "Maximum distance to known exon boundary" are additionally reported where breakpoints are not at canonical exon boundaries.
- Allow fusions with novel exon boundaries in both genes: When enabled, fusions with novel exon boundaries in both genes are reported. If not enabled, fusions with just one novel breakpoint are reported. This option is only relevant when Detect with novel exon boundaries is enabled. By default, this parameter is not enabled, to reduce the number of false positive fusions. Enabling it is useful for exhaustive searches of novel fusions.
- Only use fusion primer reads: When enabled, the input sequence list is filtered to retain reads that are annotated as originating from a primer that is designed for fusion calling. This option requires that reads are annotated with by the Biomedical Genomics Analysis tool Extract Reads Matching Primers
(see
https://resources.qiagenbioinformatics.com/manuals/biomedicalgenomicsanalysis/current/index.php?manual=Extract_Reads_Matching_Primers.html).
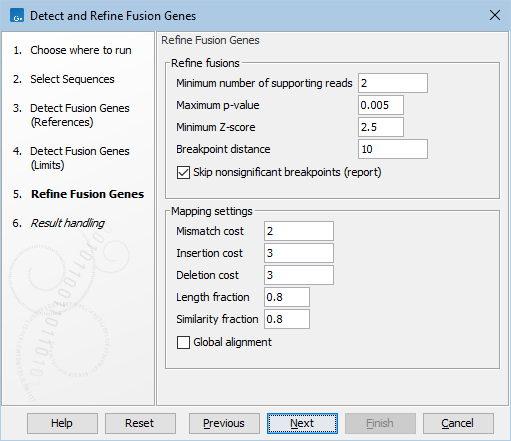
Figure 31.58: Default parameters for refining fusion genes.
- Minimum number of supporting reads: Minimum number of reads that should support a fusion. Fusions with fewer supporting reads will get a corresponding filter annotation.
- Maximum p-value: Fusions with a p-value above this threshold will get a corresponding filter annotation.
- Minimum Z-score: Fusions with a Z-score below this threshold will get a corresponding filter annotation.
- Breakpoint distance: The minimum distance from one end of a read to the breakpoint, or in other words the minimum number of nucleotides that a read must cover on each side of the breakpoint, for it to be counted as a fusion supporting read. If you set this value to 10, reads which only covers 9 bases on one side of the breakpoint will not count as fusion evidence.
- Skip nonsignificant breakpoints (report): When enabled, nonsignificant breakpoints are not added to the report.
The remaining parameters apply to the RNA-Seq read mapping to the artificial references (see Mapping settings for details).
Subsections
