Splitting a contig
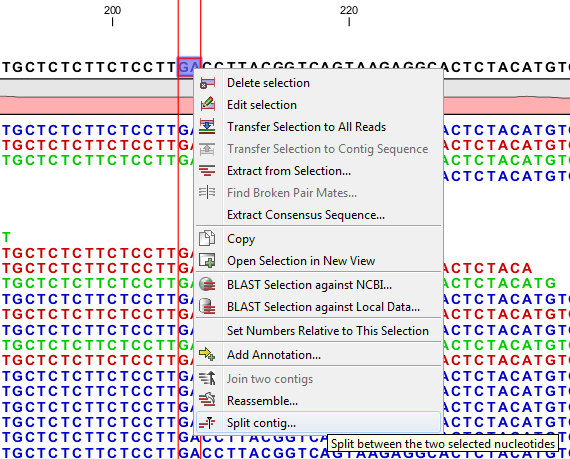
Figure 2.9: Splitting a contig
In case of misassemblies made by the de novo assembler it can be necessary to split a contig. For example, if the scaffolder has produced an erroneous scaffold or if two fragments that do not belong together have been joined into one contig, this tool can be used to split the scaffold or contig respectively. Splitting contigs is performed by selecting two nucleotides in a contig using a contig read mapping or by selecting two nucleotides in a match in the match view. After selecting two nucleotides right clicking the selection will bring up a menu where Split Contig between the two selected nucleotides can be selected (figure 2.9). This brings up a dialog where reads intersecting the split can be distributed between the resulting two contigs (figure 2.10). Click Finish to perform the split.
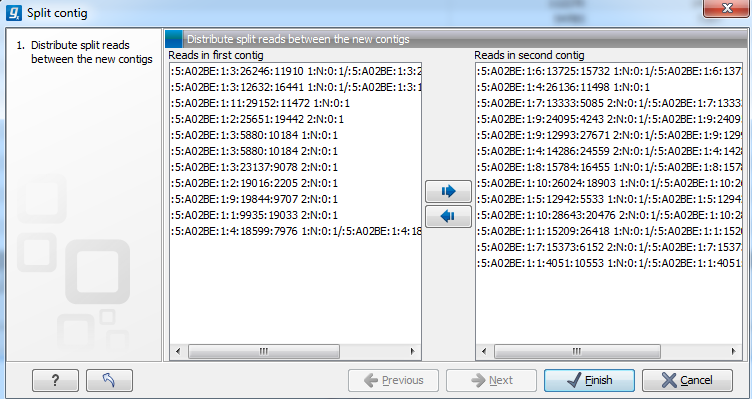
Figure 2.10: Dialog for distributing reads between split contigs
The contig will be split between the two selected nucleotides. If a contig contains reads that intersects the split region, the two contigs, which are the result of the split, will be extended with nucleotides from the other contig to preserve read alignments. As a simple example consider a split where a single read intersects the split position. If the read is placed to the left of the split, the left contig is extended with nucleotides from the right contig such that alignment of the read is preserved in the left contig. Consequently, a split at a position with intersecting reads will result in two contigs containing overlapping regions. Besides preserving the alignment of intersecting reads, the extension of split contigs is often convenient as the extended area will often overlap with the correct neighboring contig. Figure 2.11 illustrates the left half of a split where the split function has annotated the split position together with the region of the contig that overlap with the right half of the split.
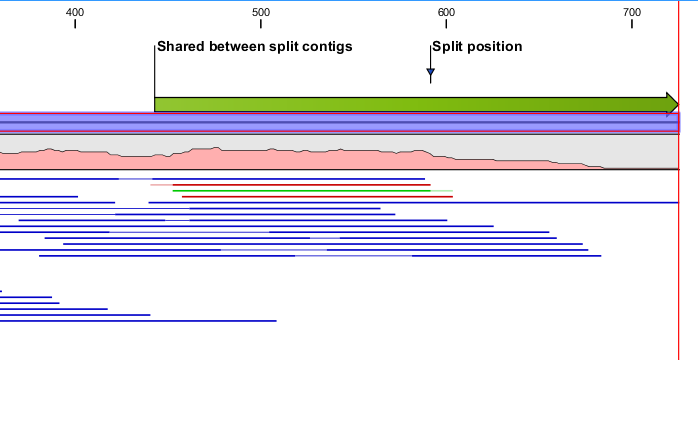
Figure 2.11: Left contig of a split where the contig shares a small region with the right contig
