How to run the Analyze Contigs tool
To run Analyze Contigs, go to:
Tools | Genome Finishing Module (![]() ) |
Analyze Contigs (
) |
Analyze Contigs (![]() )
)
This opens the dialog shown in figure 3.1.
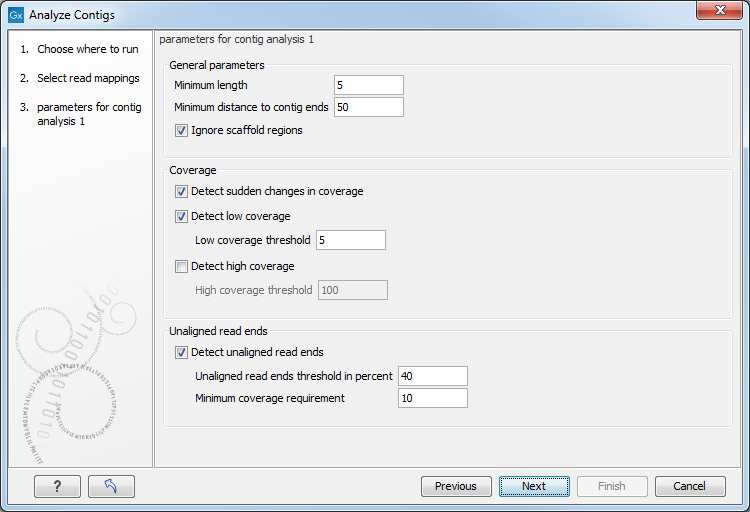
Figure 3.1: Select the contigs to be analyzed.
Select the contigs and click Next. This leads to the Set parameters for contig analysis 1 step shown in figure 3.2.
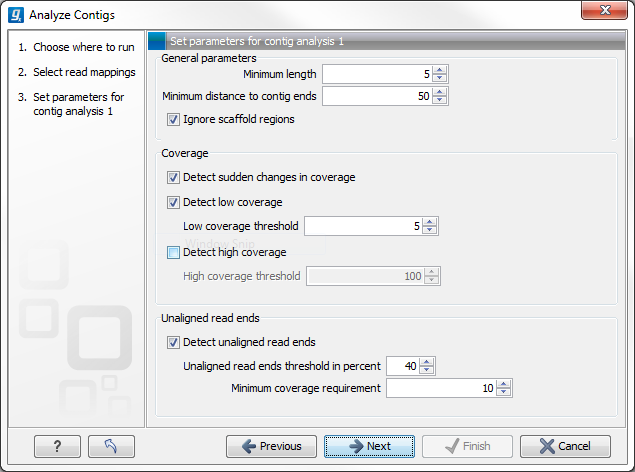
Figure 3.2: Set parameters for contig analysis 1.
The parameters to be specified in this step are:
- General parameters
- Minimum length. Specifies the minimum length of annotations. Does not apply to "sudden changes in coverage" and "unaligned ends".
- Minimum distance to contig ends. Specifies the minimum distance an annotation must have to the contig ends.
- Ignore scaffold regions. By ticking the box, regions between scaffolded contigs are ignored.
- Coverage
- Detect sudden changes in coverage. A sudden change in coverage in adjacent regions can imply a misassembly.
- Detect low coverage. Regions with low coverage can indicate a misassembly. Ticking the box allows specification of a threshold value for the minimum number of required overlapping reads.
- Detect high coverage. Regions with high coverage can indicate a misassembly. Ticking the box allows specification of a threshold value for the maximum number of accepted overlapping reads.
- Unaligned read ends
- Detect unaligned read ends. Unaligned ends of reads can imply a misassembly. Ticking the box allows specification of a threshold value for unaligned ends, which is the maximum percentage of unaligned read ends allowed at a position compared to neighboring positions.
- Minimum coverage requirement. Specifies the minimum amount of coverage required before checking for unaligned ends.
After adjustment of the parameters, click Next(figure 3.3).
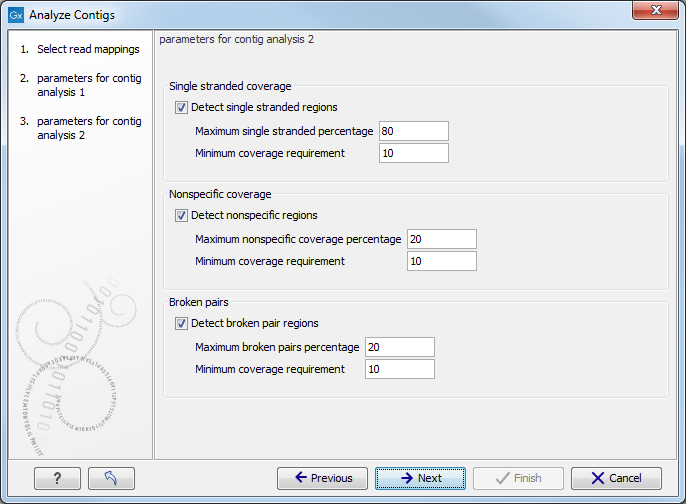
Figure 3.3: Set the parameters for contig analysis 2.
The parameters to be specified in this step are:
- Single stranded coverage
When Detect single stranded regionsis checked, regions with single stranded coverage are detected using the specified parameters:
- Max single stranded percentage specifies the maximum percentage difference between coverage of either strand with the extremes being 0% that allows only the same number of reads in both directions, and 100% that allows all reads to be in one direction. Hence, with a max single stranded percentage of 80%, single stranded regions will be detected when the difference in the number of reads in each direction exceeds 80%.
- Minimum coverage requirement. Specifies the minimum amount of coverage required before checking for single stranded coverage.
- Nonspecific coverage
When Detect nonspecific regions is checked, regions with nonspecific coverage (reads with ambiguous mapping) are detected according to the following parameters:
- Max nonspecific coverage percentage is the allowed percentage of nonspecific coverage. Only regions above this percentage are detected.
- Minimum coverage percentage is the minimum amount of coverage required before checking for nonspecific coverage.
- Broken pairs
When Detect broken pairs is checked, regions with broken pairs are detected according to the following parameters:
- Max broken pairs percentage is the allowed percentage of broken pairs.
- Minimum coverage requirement Only regions above this value are detected.
The final step shown in figure 3.4 is to specify the Output options and the Result handling.
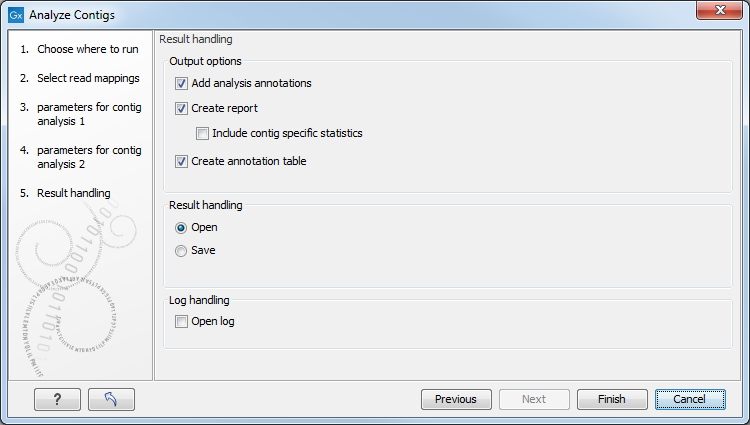
Figure 3.4: Set output parameters for contig analysis.
- Add analysis annotations. When checked, annotations are added to the regions detected in the contig analysis.
- Create report. When checked, a report is generated containing statistics on the problems identified. This report is useful for quickly evaluating the quality of an assembly.
- Include contig specific statistics. When checked, the report will contain a section for each contig with statistics for only that contig.
- Create table.
