How to run the Annotate from Reference tool
To launch Annotate from Reference, go to:
Tools | Genome Finishing Module (![]() ) |
Annotate from Reference (
) |
Annotate from Reference (![]() )
)
This opens the dialog shown in figure 13.4 where at least one Align Contigs result must be selected. Click Next.
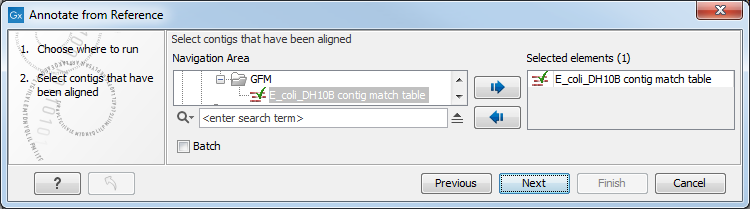
Figure 13.4: Select Align Contigs results.
The next step in figure 13.5 allows you specify the following:
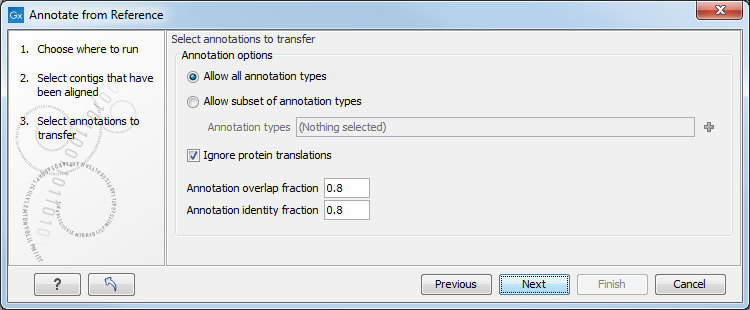
Figure 13.5: Specify parameters for deciding in which cases an annotation should be transferred.
- Allow all annotations types found on the input reference sequences to be transferred.
- Allow subset of annotations types will transfer only a subset of all annotations found on the input reference sequences. You can choose from a pop up menu which subset you would like to transfer.
- Ignore protein translations. If this option is selected, any annotation containing "translation" as qualifier text will not be transferred to the new sequence, since it would reflect the translation of the original sequence.
- Annotation overlap fraction gives the fraction of the annotation length that must be included in the contig to to transfer annotations.
- Annotation identity fraction gives the fraction of matching nucleotides between contigs and annotated reference regions needed to transfer annotations.
Figure 13.6 shows the output options. They include generation of reports and tables containing information on the annotations that were transferred and those that were not. You can also add annotations to aligned contigs, and create contigs with annotations. Click Finish to transfer annotations.
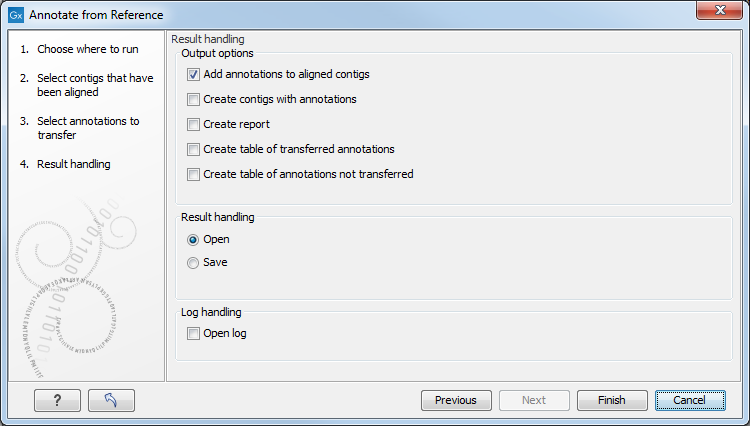
Figure 13.6: Output options for the Annotate from Reference tool.
