Taxonomic Profiling
The Taxonomic Profiling tool is designed to determine which known organisms are in a whole shotgun metagenomic sample, and how abundant they are. To this end, the tool will map each input read to a reference database of complete genomes. If a host organism genome is provided, the mapping phase will disregard reads that it deems to have originated from the host. Paired reads that cannot map as an intact pair will also be dismissed. After mapping, the tool performs qualification (by assigning the read to a taxon in the database if a match is found) and quantification of the abundance of each qualified species, and finally compile the results in an abundance table.
The purpose of the qualification phase is to determine whether a particular reference sequence is represented in the sample. We do this by observing that, if a given reference is present in the sample, then it is highly likely that the reference is evenly covered across the entire reference. Thus, we disqualify any reference that is not evenly covered across the entire reference.
The purpose of the quantification phase is to estimate how abundant the qualified references are. This is done in two different ways depending on whether reads are mapping uniquely to a given reference or not. If a given qualified reference contains reads that map uniquely to that reference and no other references in the database, then abundance for this reference is estimated from the peaks of uniquely mapping reads (reads mapping uniquely to a specific genome in a large database tend to do so uniformly within clusters and thus forming peaks). If a given qualified reference contains no uniquely mapping reads, then abundance is estimated from the total amount of nucleotides mapping to the reference and the total length of the reference. The final step of quantification is to coalesce patterns of disqualified coverage onto higher-level taxons.
Note that the Taxonomic Profiling tool is currently optimised for specificity and as a consequence very conservative in the detection of low abundance species in samples of average coverage: for example, to have a 50% chance to detect a species, its coverage needs to be at at least 6.6. This stringency limitation will be addressed in the future releases of the software.
To run the Taxonomic Profiling tool, go to Microbial Genomics Module (![]() ) | Metagenomics (
) | Metagenomics (![]() ) | Taxonomic Analysis (
) | Taxonomic Analysis (![]() ) | Taxonomic Profiling (
) | Taxonomic Profiling (![]() ).
).
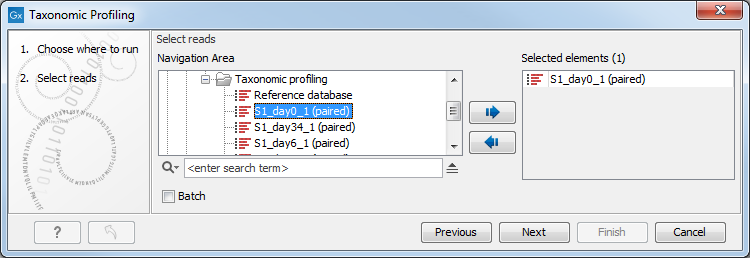
You can select one or several read files to analyze (figure 5.1). When choosing several read files, they will be considered as belonging to one single sample unless the batch mode option is checked, in which case each file will be considered as an individual sample.
In the "Parameters" dialog , provide the reference database you will use to map the reads (figure 5.2). Note that the first time you run the tool with a given database, the analysis will take longer because it is indexing and caching the database as indicated by the warning message in the dialog. Analysis time will be improved in subsequent runs, when the workbench is able to use the index generated the first time around.
You can also choose to "Filter host reads". You must then specify the host genome (for example the Homo sapiens hg19 in the case of human microbiota). Finally, checking the option "Auto-detect paired distances" will generate an estimate of the paired distance range in an additional section of the report output by the tool.
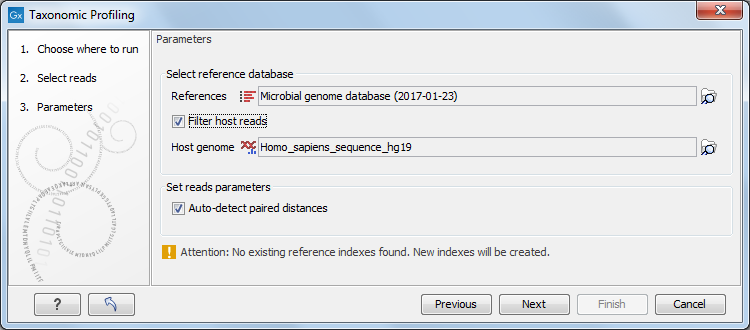
Figure 5.2: Select the reference, and potentially choose to filter against an host genome to remove potential contamination.
Finally, choose from the different output options and Open or Save the results (figure 5.3).
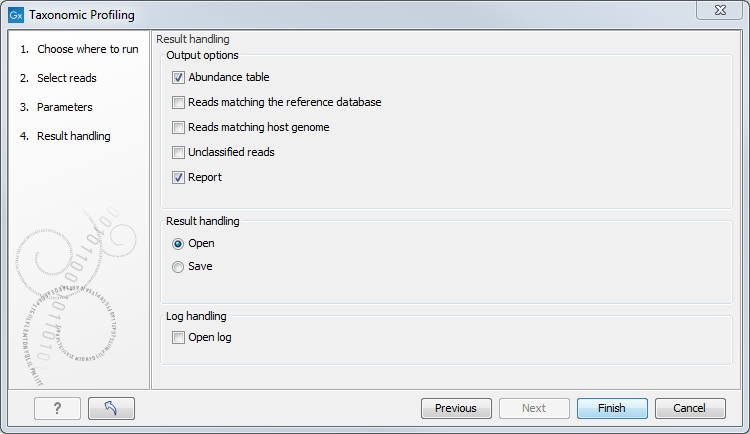
Figure 5.3: Workflow output options.
The tool will generate by default an abundance table as well as a report with a list of the taxons and their abundances. You can choose to output additional files such as a sequence lists of the reads matching the reference database and those matching the host, as well as the unclassified reads.
Subsections