OTU clustering tool outputs
The OTU clustering tool produces several outputs: a sequence list of the OTU centroids, and two abundance tables with the newly created OTUs or the chimeras. Each table give abundance of the OTU or chimeras at each site as well as the total abundance for all samples. Note that if the input contains paired-end sequences, each pair is counted as one read.
In the OTU merge report - generated if the input reads were paired - the following statistics are given for each sequence list input in the tool: a summary of merged and non merged sequences, an alignment score distribution and a merged pairs length distribution.
The tool also produces a report (figure 4.4).
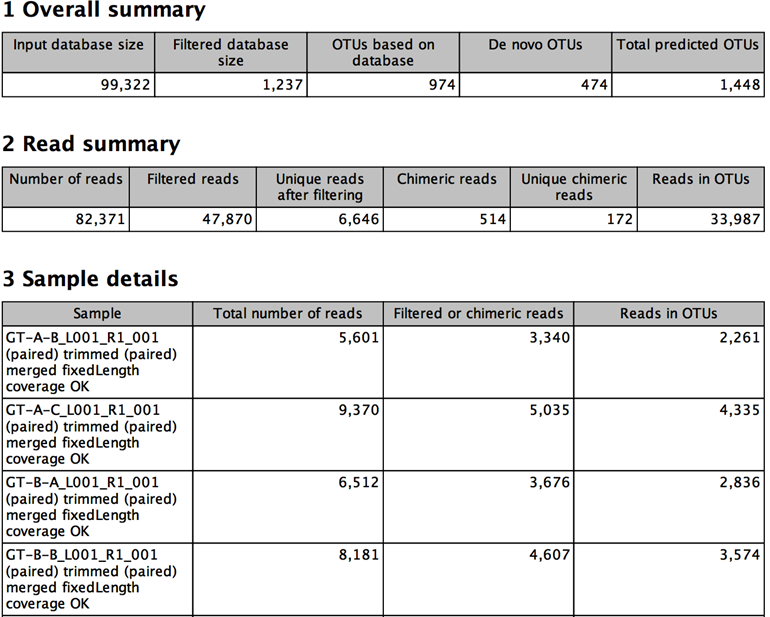
Figure 4.4:
Example of report produced by the OTU clustering tool.
The content of the report is stated below.
- In the section Overall summary
- Input database size The number of sequences in the input OTU database.
- Filtered database size The number of sequences in the input OTU database having input reads mapped to it.
- OTUs based on database The number of OTUs based on a sequence from the database.
- De novo OTUs The number of OTUs not based on a sequence from the database.
- Total predicted OTUs The total number of OTUs found.
- In the section Read summary
- Number of reads The number of input reads
- Filtered reads The number of reads filtered due to the minimum occurrences parameter.
- Unique reads after filtering The number of unique reads after filtering. This is the number of candidates for OTUs before clustering.
- Chimeric reads The number of reads detected as chimeric during clustering.
- Unique chimeric reads The number of unique reads detected as chimeric.
- Reads in OTUs The number of reads that contribute to the output OTUs.
- In the section Sample details
- Sample The name of the sample for which the following details are shown.
- Total number of reads The number of input reads from the given sample.
- Filtered or chimeric reads The number of reads from the given sample that were filtered due to the minimum occurrences parameter or detected as chimeric during clustering.
- Reads in OTUs The number of reads from the given sample that contribute to the output OTUs.
