Create Microbial Reference Database
The Create Microbial Reference Database tool downloads selected references from GenBank and RefSeq, and outputs a single sequence list with all the necessary annotations for the taxonomic profiling (i.e., assembly IDs).
To run the tool, go to:
Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Create Microbial Reference Database
) | Create Microbial Reference Database
In the first window (figure 15.1), select the source of the database you wish to generate.
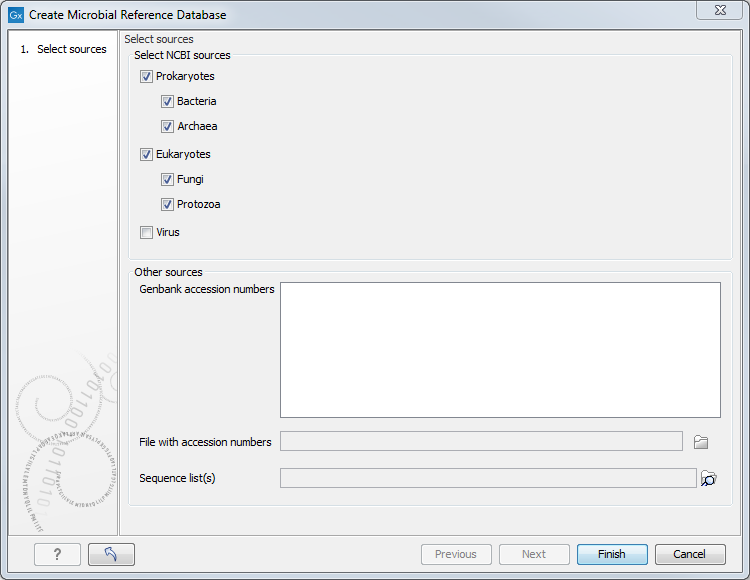
Figure 15.1: Select the references you want to download.
You can choose from:
- NCBI sources
- Prokaryotes: Bacteria and/or Archaea
- Eukaryotes: Fungi and/or Protozoa
- Virus. Note that downloading choosing this option will result in both virus and bacterial assemblies. Indeed, viruses are identified according to their BioProject ID, but this ID also refers to bacterial assemblies that were sequenced together with the virus. Filtering the table on taxonomy will allow you to only see viruses.
- Other sources
- Provide a list of Genbank accession numbers in the white field, or
- Browse your computer for a file with accession numbers, or
- Browse the Navigation Area of the workbench for a sequence list. The corresponding references will be appended to the downloaded sequence list automatically.
The time it will take to download the data (such as assembly summaries, genome report) depends on how many databases are downloaded and the bandwidth of your internet connection. No sequence data is downloaded at this point.
The output is a table as in figure 15.2.
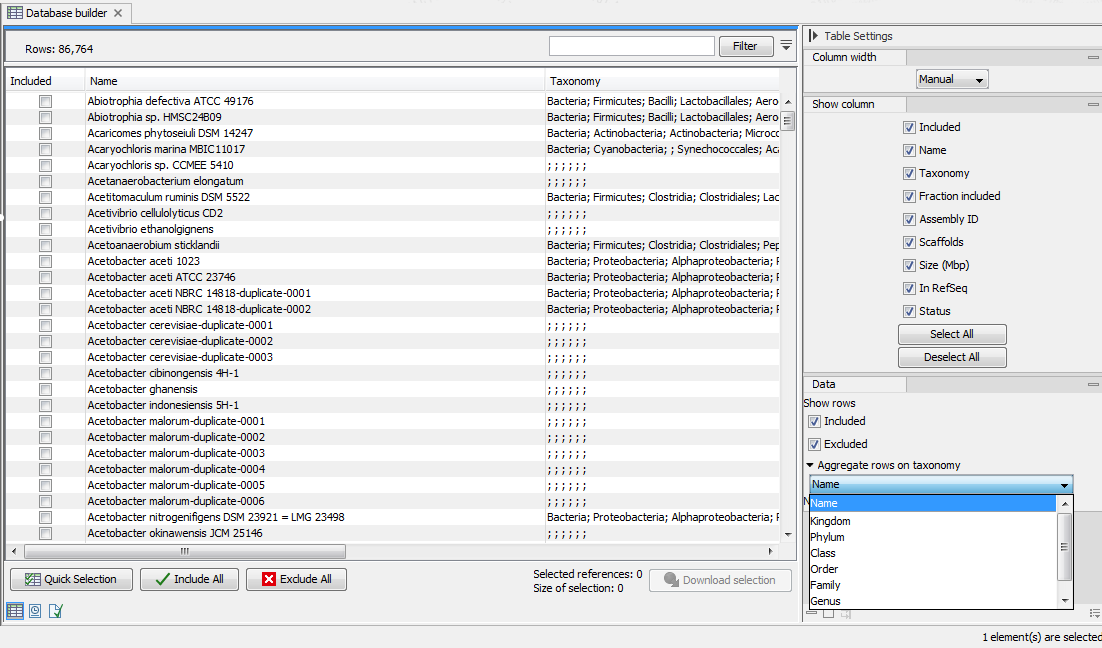
Figure 15.2: Output table from the Create Microbial Reference Database tool.
The resulting table can be used to design your own database. A series of functionality can help you filter and sort the table to extract the information relevant to your project.
- Use the "Quick selection" button to quickly select predefined subsets for
download:
- Single scaffold complete genomes in RefSeq
- Complete genomes in RefSeq
- All complete genomes
- Aggregate the table to a specified taxonomic group using the drop down menu in the "Data" palette of the side panel. Use the category "Name" to de-aggregate the table.
- Use filter(s) to keep only the rows you are interested in, and click on the button "Include all" to create a database with the remaining rows.
- Alternatively, click on "Include all" rows first, set one or several filters, and use the button "Exclude all" on the remaining rows. Clear the filter(s) by clicking on the red buttons next to each filter set. The rows not filtered away in the second step should still be checked.
Once the table contains all desired rows, click "Download selection". Close to the button, you can check how many references are selected, and an estimate of the total size of the selection.
The dialog shown in figure 15.3 allows you to set an additional filter "Minimum contig length". It also warns about the memory and disk requirements that will be needed to later run the Taxonomic Profiling tool with the database you are about to download.
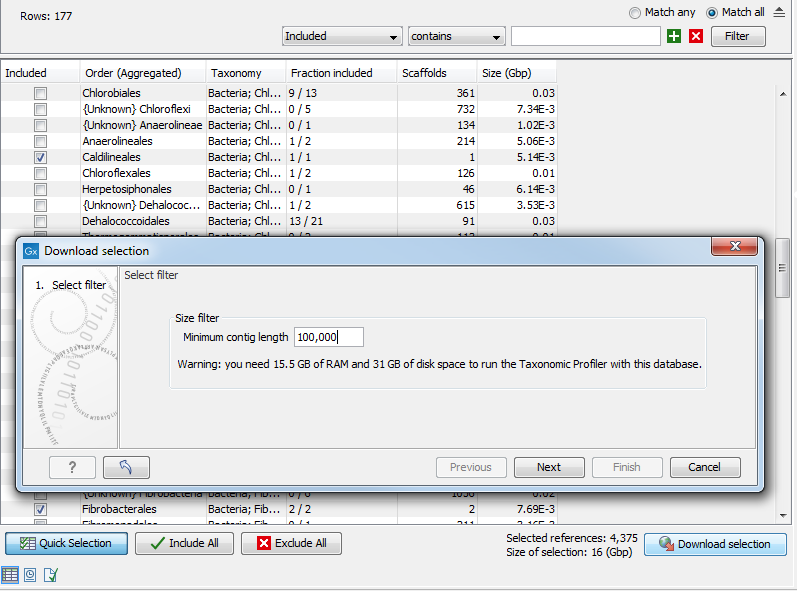
Figure 15.3: The "Download selection" wizard.
