Metadata
Metadata refers to information about data. In the context of the CLC Genomics Workbench, this usually means information about samples. For example a set of reads could come from a particular specimen at a particular time point with particular characteristics. The specimen, time and characteristics would be metadata for that set of reads.
Examples in this chapter refer to tools present in the CLC Genomics Workbench, but the principles apply to other CLC Workbenches.
What is metadata used for?
Core uses of metadata in CLC software are listed below, along with a reference for where further information on that aspect can be found: Running tools where characteristics of the data elements are relevant. Examples are the differential expression tools, described in Differential Expression.- Defining batch units when launching workflows or tools in batch mode, and for launching workflows in batches where more than one input should be changed for each batch run, described in Launching workflows individually and in batches.
- Distributing data to the relevant input channels in a workflow when using Collect and Distribute elements, described in Batching part of a workflow.
- Finding and selecting data elements associated with the metadata. Workflow result metadata tables are of particular use when reviewing results generated by workflows run in batch mode and are described in Workflow outputs and workflow result metadata tables.
Metadata tables
An example of a metadata table in the CLC Genomics Workbench is shown in figure 10.1. Each column represents a property of a sample (e.g., identifier, height, age, treatment) and each row contains information relevant to a sample. One column will be designated the key column. That column must contain unique entries and is used when associating data elements with a metadata row.
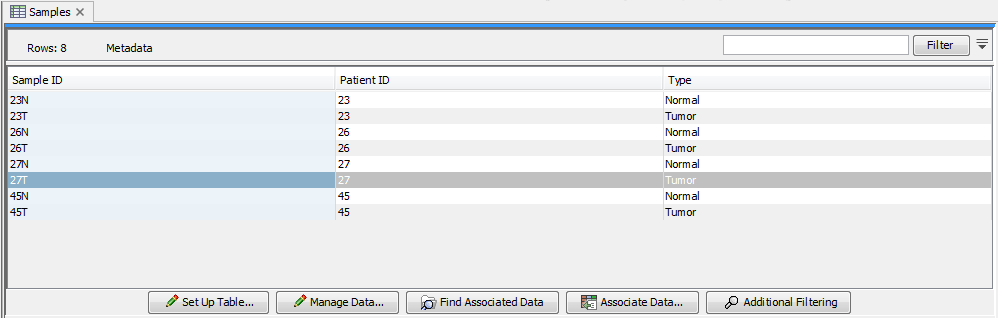
Figure 10.1: A simple metadata table, with the key column highlighted in blue.
A row in a metadata table can be associated with one or more data elements, such as sequence lists, expression tracks, variant tracks, etc. Associating data elements with relevant metadata rows, automatically or manually, is covered in section Associating data elements with metadata
The most common way to create a metadata table is to import an Excel format file, as described in Importing metadata. Metadata tables can also be generated by workflows as described in Workflow outputs and workflow result metadata tables
Searching for metadata tables based on their contents is described in Quick search.
Metadata Elements table
The data elements associated particular metadata rows can be listed by selecting the metadata rows of interest and clicking on the Find Associated Data button. This opens the Metadata Elements table, where the associated data will be listed, as shown in figure 10.2.
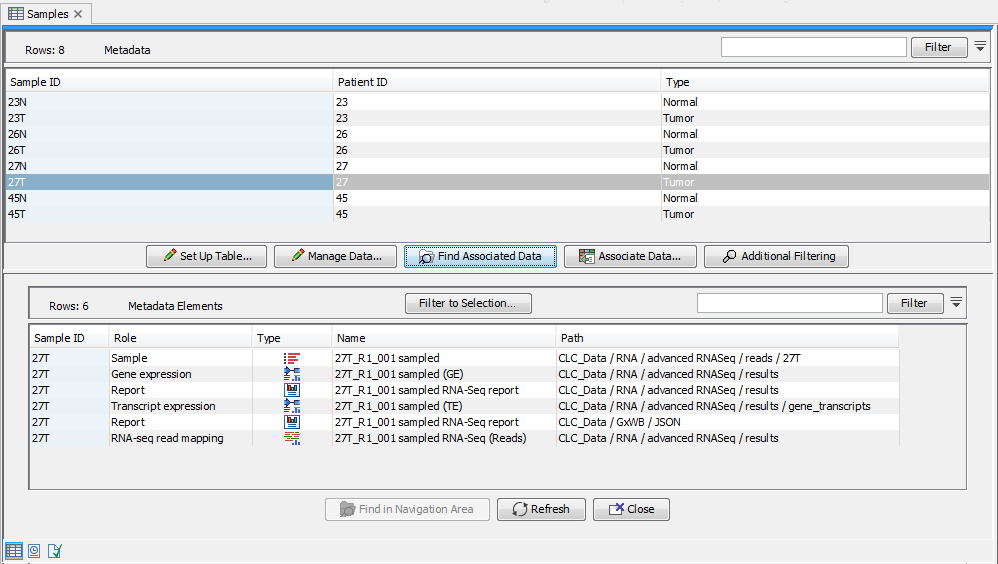
Figure 10.2: A Metadata Table and corresponding Metadata Elements table showing elements associated with sample 27T.
When a data element is associated with a metadata row, the outputs of analyses involving that data often inherit the metadata association automatically. This means that a given row in a metadata table can be associated with several data elements. For example, at first a sample might be associated with a sequence list, but after analysis, the same metadata row could be associated with various additional elements in the Metadata Elements table, can be seen in figure 10.2.
Inheritance of metadata associations requires that a single association can be unambiguously identified for an output when a tool is run. If an output is derived from two ore more inputs with different metadata associations, then no association will be inherited.
Subsections
- Creating metadata tables
- Associating data elements with metadata
- Working with data and metadata
- Moving, copying and exporting metadata
- Editing Metadata tables
