Mapping settings
When the reference has been defined, click Next and you are presented with the dialog shown in figure 30.5.
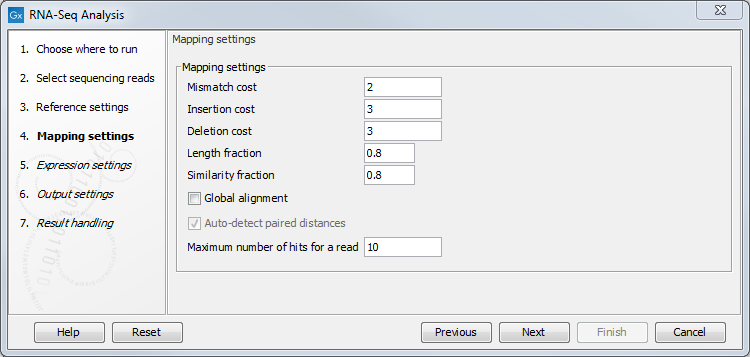
Figure 30.5: Defining mapping parameters for RNA-Seq.
The mapping parameters are identical to those applying to Map Reads to Reference, as the underlying mapping is performed in the same way. For a description of the parameters, please see Mapping parameters.
For the estimation of paired reads distances, RNA-Seq uses the transcript level reference sequence information. This means that introns are not included in the distance measurement. The paired distance measurement will only include transcript sequence, reflecting the true nature of the sequence on which the paired reads were produced.
In addition to the generic mapping parameters, two RNA-Seq specific parameters can be set:
- Maximum number of hits for a read. A read that matches equally well to more distinct places in the reference sequence than the 'Maximum number of hits for a read' specified will not be mapped. If a read matches to multiple distinct places, but less than or equal to the specified maximum number, it will be assigned to one of these places by the EM algorithm (see EM estimation algorithm). Note that to favor accurate expression analysis, it is recommended to have this value set to 10 or more.
Concept of hits and distinct places in the reference
The definition of a distinct place in the reference sequence is complicated. We are describing here example cases where the option "Genome annotated with genes and transcripts" is selected in the previous "Reference settings" step, meaning that reads are aligned to genes and transcripts.
- In an example case where 2 genes are overlapping, a read will count as one hit because it corresponds to the same reference sequence location. This read will be assigned to one of the genes by the EM algorithm.
- In an example case where a gene has 10 transcripts and 11 exons, and all transcripts have exon 1 plus one of the exons 2 to 11. Exon 1 is thus represented 11 times in the references (once for the gene region and once for each of the 10 transcripts). Reads that match to exon 1 will thus match to 11 of the extracted references. However, when the mappings are considered in the coordinates of the main reference genome, it becomes evident that the 11 match places are not distinct but in fact identical. In this case this will just count as one hit.
- In a more complicated example, a gene has different splicing, for example transcripts with longer versions of an exon than the others. In this case you may have reads that may either be mapped entirely within the long version of the exon, or across the exon-exon boundary of one of the transcripts with the short version of the exon. These reads are ambiguously mapped (they appear in yellow in a track view), and count as as many hits as different ways they map to the reference. Setting the 'Maximum number of hits for a read' parameter too low could leave these reads unmapped, eliminating the evidence for the expression of the gene to which they mapped.
