Importing and viewing trace data
A number of different binary trace data formats can be imported into the program, including Standard Chromatogram Format (.SCF), ABI sequencer data files (.ABI and .AB1), PHRED output files (.PHD) and PHRAP output files (.ACE) (see Bioinformatic data formats).
After import, the sequence reads and their trace data are saved as DNA sequences. This means that all analyses that apply to DNA sequences can be performed on the sequence reads.
You can see additional information about the quality of the traces by holding the mouse cursor on the imported sequence. This will display a tool tip as shown in figure 19.1.

Figure 19.1: A tooltip displaying information about the quality of the chromatogram.
The qualities are based on the phred scoring system, with scores below 19 counted as low quality, scores between 20 and 39 counted as medium quality, and those 40 and above counted as high quality.
If the trace file does not contain information about quality, only the sequence length will be shown.
To view the trace data, open the sequence read in a standard
sequence view (![]() ).
).
The traces can be scaled by dragging the trace vertically as shown in figure figure 19.2. The Workbench automatically adjust the height of the traces to be readable, but if the trace height varies a lot, this manual scaling is very useful.
The height of the area available for showing traces can be adjusted in the Side Panel as described inTrace data settings in the Side Panel.
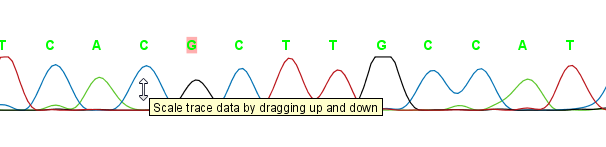
Figure 19.2: Grab the traces to scale.
Subsections
