Download Genomes
In the Download Genomes tab, you can access genomes and associated genomic data such as annotations and known variants (figure 8.4). The data is not provided or hosted by QIAGEN. The workbench only provides an easy way to retrieve data that should otherwise have been downloaded and imported.
The list of organisms is dynamically updated by QIAGEN independently of Workbench versions, so you will always see the most recent list of organisms. Select the organism of your choice to see what is available for download, and check the elements you want to include in the downloading process (the size of the download file is updated with each selected element). Please note that the file size displayed in the setup window for the Download Genomes tool refers to the size of the compressed text files, which the tool is retrieving from the provider's depository. The size of the track objects will be, after decompression and conversion from text to the .clc track format, larger.
The reference sequence will be downloaded automatically from Ensembl. You can also choose to select an existing genome sequence track from your Navigation Area to initiate the download. In that case, the reference sequence has to match the genome definition built into the download tool. This means that the name and length of the chromosomes in your reference sequence have to match the genome definition of the tool.
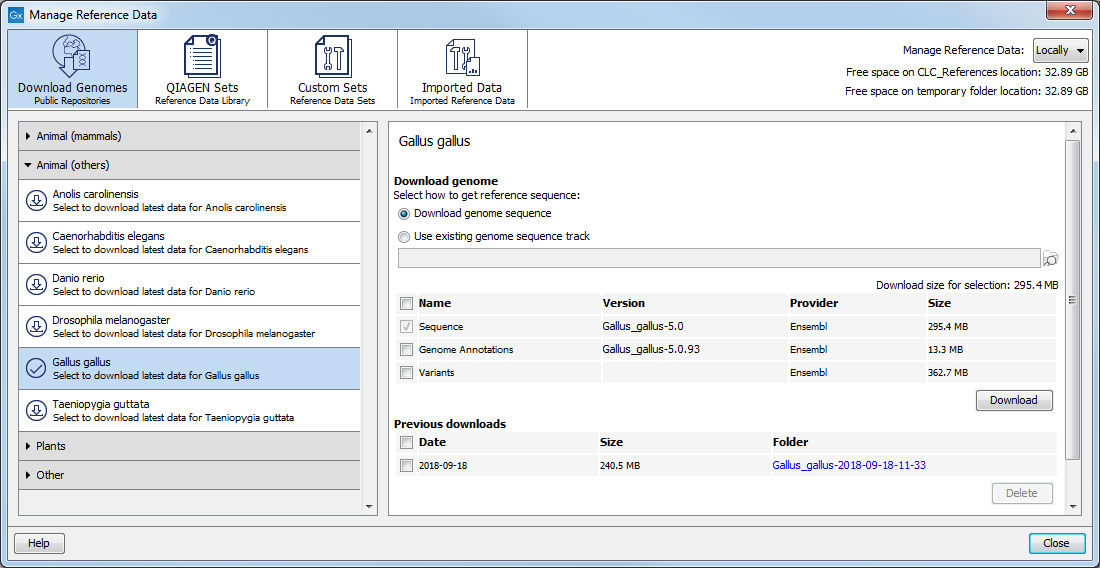
Figure 8.4: Download genomes and associated data for selected organisms.
All data downloaded with this tool will be tracks (either sequence tracks or various kinds of annotation tracks).
Which types of annotation are available for download is different from organism to organism and depends on the data sources that QIAGEN has included for download.
The Ensembl human gene annotation download will produce various tracks describing the chromosomal positions of features such as exons, genes, untranslated region (UTRs), transcripts, selenocysteines, coding sequences (CDSs) and mRNAs. The tracks will be saved in a folder called "Genomes" in the CLC_References folder of the Navigation Area. Previous downloads are listed in the Reference Data Manager dialog, and can be deleted from there if needed (but note that this requires admin rights if the files are located on a server).
When GFF3 files are imported, an output track will be issued for each feature type present in the file (see here), and in addition, the Workbench will generate an (RNA) track that aggregates all the types that were "RNA" into one track (i.e., all the children of "mature_transcript", which is the parent of "mRNA", which is the parent of the "NSD_transcript"); and a (Gene) track that includes genes and Gene-like types annotations like ncRNA_gene, plastid_gene, and tRNA_gene. These "(RNA)" and "(Gene)" tracks are different from the ones ending with "_mRNA" and in "_Gene" in that they compile all relevant annotations in a single track, making them the track of choice for subsequent analysis (RNA-Seq for example).
UCSC Genome Browser available data include dbsnp variants and chromosome ideograms, also called a cytogenetic ideograms, i.e., a chromosome map with numbered banding patterns that shows the relationship between the two chromosome arms and the centromere (figure 8.5). However, variants downloaded from UCSC will not be annotated on the mitochondrial genome.
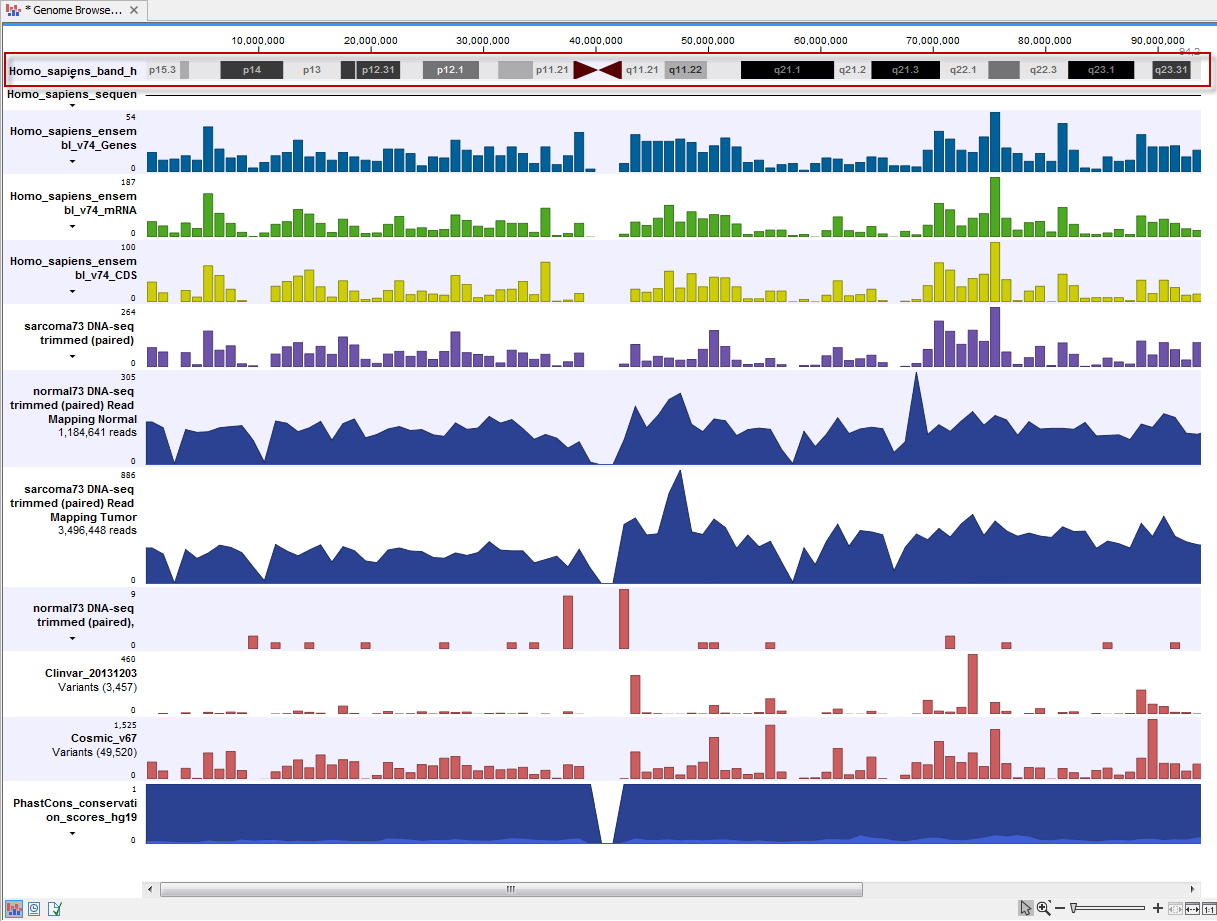
Figure 8.5: The ideogram is particularly useful when used in combination with other tracks in a track list. In this figure the ideogram is highlighted with a red box.
