Assembly Viewer
The assembly viewer program shows assemblies in a text based terminal window. It is useful for getting a quick overview of the data and for investigating interesting places.
The program takes one or more assembly files as parameters. For large assemblies, it may take a little while to start since the reads have to be sorted for viewing. The key bindings are as follows:
| Key | Description |
| Arrows | Move view. |
| 0-9 | Any (possibly multi digit) number followed by any other key: move to that position. Follow by `K' to multiply by 1,000, or `M' to multiply by a million. |
| z | Center vertical position on reads. |
| v | Scroll left to interesting part and center horizontally. |
| b | Scroll right to interesting part and center horizontally. |
| c | Toggle color scheme. |
| m | Toggle position marks. |
| e | Toggle how to show unaligned ends. |
| r | Toggle between contigs. |
| j | Toggle joint read view. |
| p | Move to same position as for last contig. |
| h | Show help screen. |
| s | Search for a sequence in the reference. |
| q | Quit. |
Using shift together with one of the toggle keys (`C', `E', `R' and `M') cycles the other direction. Using shift with one of the movement keys (including arrows) makes the movement faster. This also applies to the `K' and `M' keys for sequence positions. Figures 6.1-6.4 show some screen shots and examples.
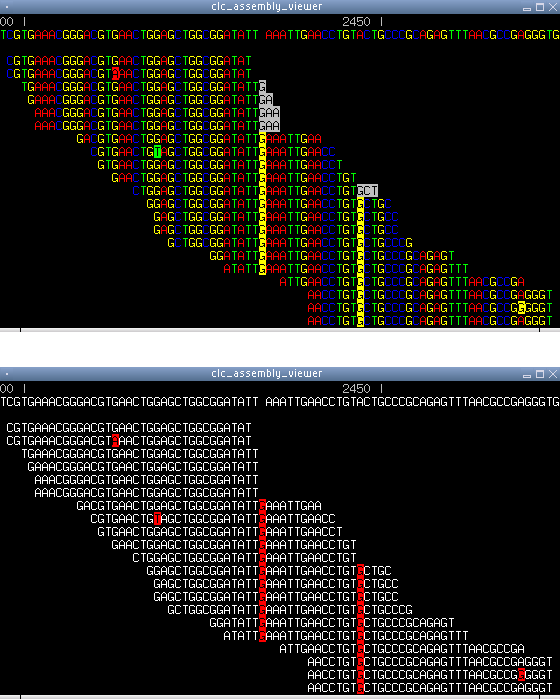
Figure 6.18: Two screen shots from the assembly viewer. Top) Residue
coloring. Residues differing from the reference are highlighted. The
first column of highlighted G's is an insertion, the second is a
mutation (the reference residue is A in that position). The reversed
gray residues at the end of some of the reads are not
aligned. Bottom) Another color scheme, where differences are easier
to spot. Here the unaligned residues have also been turned off.
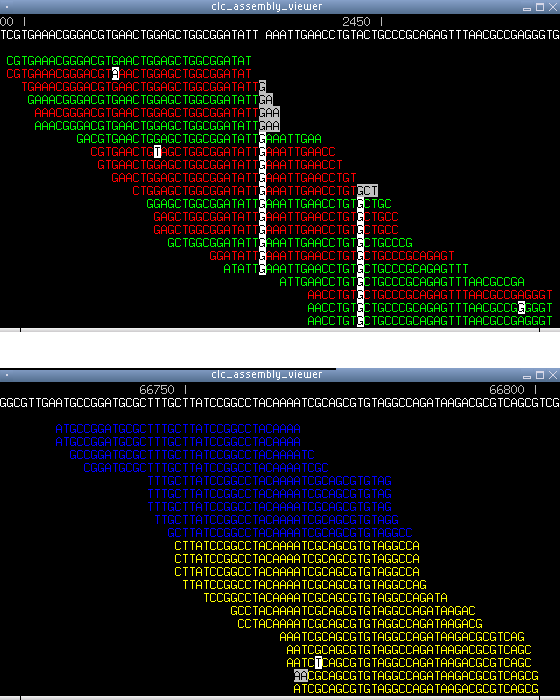
Figure 6.19: Another screen shot from the assembly viewer. Top) reads are colored according to the direction. Green is forward, red is reverse. Bottom) a yellow color indicate reads that map uniquely while a blue color indicate reads which map ambiguously, i.e. they map with the same score at multiple positions which often indicate a repetitive region.
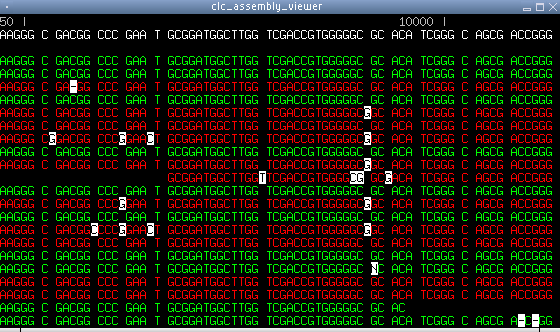
Figure 6.20: A screen shot with 454 sequencing data. The
directional color scheme is useful for recognizing a particular type
of sequencing error with the 454 technology. Notice the position
with five inserted G's. They are sequencing errors arising from the
stretch of five G's to their left, before the C. These errors tend
to occur before a stretch of identical residues, which is why they
are only seen in the reverse reads in this case.
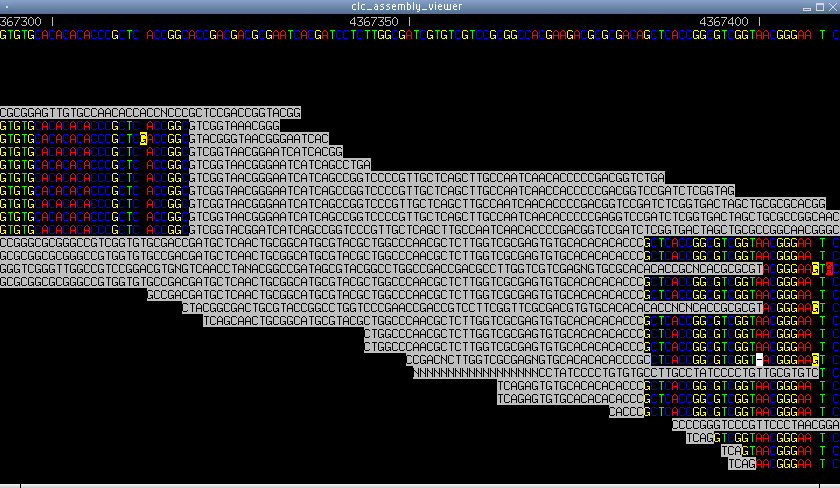
Figure 6.21: A screen shot with 454 sequencing data. This is how
a genomic rearrangement looks in a reference assembly. Suddenly the
reads do not match any more, and later another set of reads abruptly
start matching. These reads may actually be very distant in the real
genome (as opposed to the reference).
