Restriction Site Analysis
Besides the dynamic restriction sites, you can do a more elaborate
restriction map analysis with more output formats using the tool:
Toolbox | Restriction Site Table (![]() ) | Restriction Site Analysis (
) | Restriction Site Analysis (![]() )
)
You first specify which sequence should be used for the analysis. Then define which enzymes to use as basis for finding restriction sites on the sequence (see Manage enzymes).
In the next dialog, you can use the checkboxes so that the output of the restriction map analysis only include restriction enzymes which cut the sequence a specific number of times (figure 11.8).
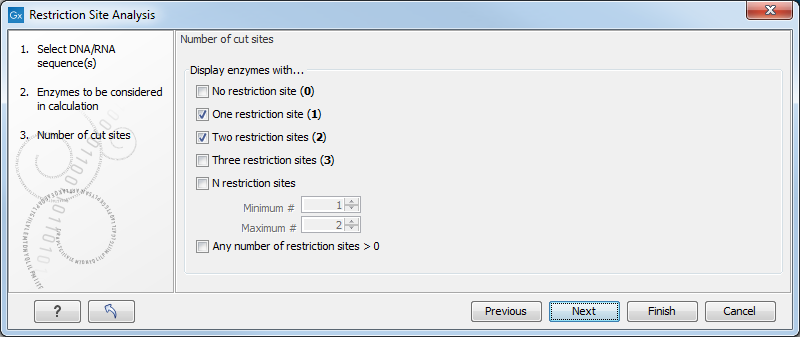
Figure 11.8: Selecting number of cut sites.
The default setting is to include the enzymes which cut the sequence one or two times, but you can use the checkboxes to perform very specific searches for restriction sites: e.g. if you wish to find enzymes which do not cut the sequence, or enzymes cutting exactly twice.
The Result handling dialog (figure 11.9) lets you specify how the result of the restriction map analysis should be presented.
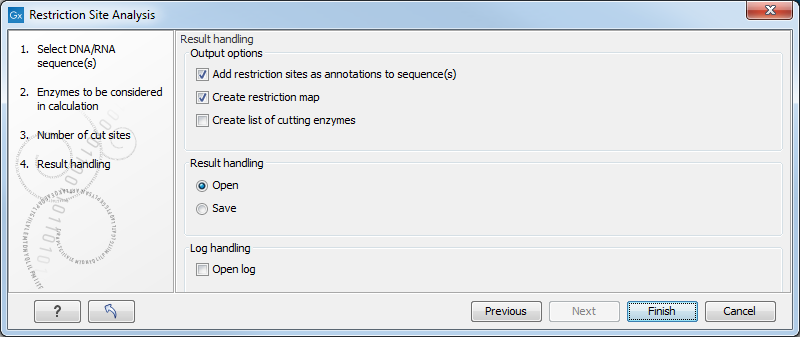
Figure 11.9: Choosing to add restriction sites as annotations or creating a restriction map.
Add restriction sites as annotations to sequence(s)
. This option makes it possible to see the restriction sites on the sequence (see figure 11.10) and save the annotations for later use.
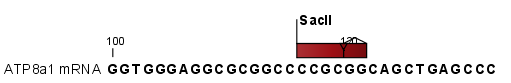
Figure 11.10: The result of the restriction analysis shown as annotations.
Create restriction map
. The restriction map is a table of restriction sites as shown in figure 11.11. If more than one sequence were selected, the table will include the restriction sites of all the sequences. This makes it easy to compare the result of the restriction map analysis for two sequences (or more).
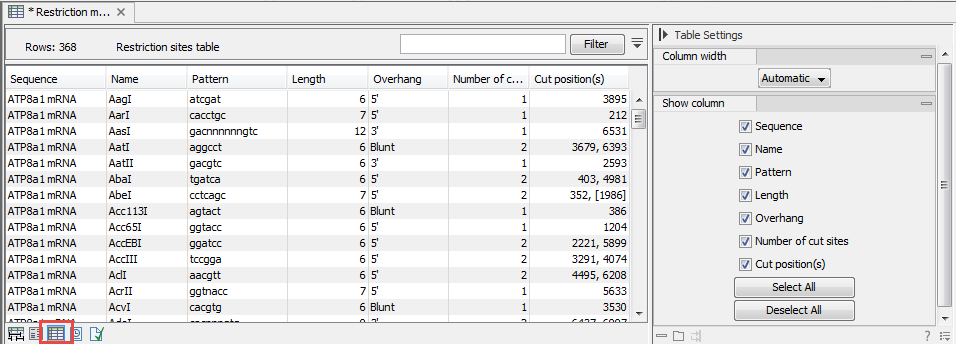
Figure 11.11: The result of the restriction analysis shown as annotations.
Each row in the table represents a restriction enzyme. The following
information is available for each enzyme:
- Sequence. The name of the sequence which is relevant if you have performed restriction map analysis on more than one sequence.
- Name. The name of the enzyme.
- Pattern. The recognition sequence of the enzyme.
- Overhang. The overhang produced by cutting with the enzyme (3', 5' or Blunt).
- Number of cut sites.
- Cut position(s). The position of each cut.
- , If the enzyme cuts more than once, the positions are separated by commas.
- [] If the enzyme's recognition sequence is on the negative strand, the cut position is put in brackets (as the enzyme TsoI in figure 11.11 whose cut position is [134]).
- () Some enzymes cut the sequence twice for each recognition site, and in this case the two cut positions are surrounded by parentheses.
