Launching workflows with Iterate elements
The section focuses on launching workflows that contain Iterate elements using the CLC Server Command Line Tools. Iterate elements are a type of control flow element, controlling the flow of data through an analysis. Iterate elements are placed at the top of a branch of a workflow that should be run multiple times, using different inputs in each run. The sets of data to use in each run are referred to as "batch units".
Collect and Distribute elements are, optionally, placed downstream of an Iterate element, where they collect outputs from the upstream iteration block and distribute them as inputs to downstream analyses. Most Collect and Distribute elements have a single input channel and a single output channel and do not require any parameters to be specified on the command line. Writing commands for other situations is described at the end of this section.
For general information about control flow elements, see the CLC Genomics Workbench manual.
The steps between an Iterate element and a Collect and Distribute element are referred to as an "iteration block". The workflow in figure 8.3 contains a single iteration block (shaded in turquoise), where steps within that block are run once per batch unit. The Collect and Distribute element (renamed to Collect Expressions) collects all the results from the iteration block and sends it as input to the next stage of the analysis (shaded in purple).
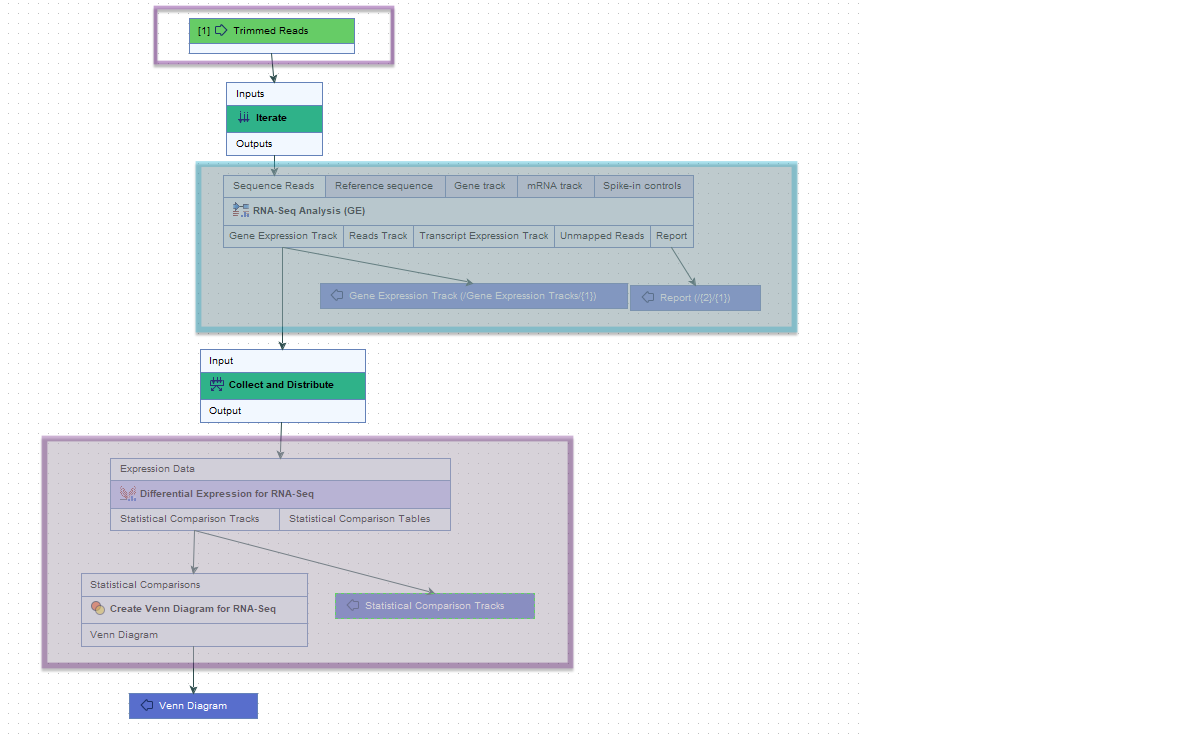
Figure 8.3: The roles of the Iterate and Collect and Distribute control flow elements are highlighted in the context of RNA-Seq and differential expression analyses.
RNA-Seq Analysis lies downstream of an Iterate element, within an iteration block (shaded in turquoise). It will thus be run once per batch unit.
Differential Expression for RNA-Seq lies immediately downstream of a Collect and Distribute element (renamed to Collect Expressions), and is sent all the expression results from the iteration block as input for a single analysis.
The following are key to launching workflows containing an Iterate element:
- Specifying how batch units are defined: by the organization of the input data or using metadata.
- For batch units specified using metadata:
- Indicating whether the metadata in a CLC Metadata Table or in an external file, specifically an Excel, CSV or TSV format file, and
- Specifying the metadata column defining the grouping of the data.
For workflows with a single Iterate element that has a single input channel and a single output channel, and where the batch units are based on the organization of the input data, no parameters relating to the Iterate element need to be provided in the command. In other cases, the parameters below need to be specified. Parameter names start with the workflow element name, which in this case was the default name, Iterate.
--iterate-iterate-unitsUsed to specify how the batch units are defined:SIMPLE(default) Based on the organization of the input data.METADATABased on metadata that will be provided.Note that when launching a workflow containing a tool requiring metadata as input, for example Differential Expression for RNA-Seq, batch units must be specified using metadata. The metadata provided to define the batch units is also used in the analysis step(s) requiring metadata.
--iterate-metadata-sourcesRequired when batch units are defined using metadata to specify how the metadata will be provided.TABLE_OBJECTIn a CLC Metadata TableFILEIn an Excel, CSV or TSV file
---iterate-metadata-tableWhenTABLE_OBJECTis defined as the metadata source, specify this parameter and provide a CLC Object URL for the CLC Metadata Table to use as the value.--iterate-metadata-fileWhenFILEis defined as the metadata source, specify this parameter and provide the location of the Excel, TSV or CSV format file to use as the value.--iterate-metadata-table-columnsThe metadata column containing the information for grouping the data into batch units. One parameter-value pair is expected per input channel in the Iterate element.
In cases where the Iterate element has multiple input channels, the first input channel is considered the primary input channel by default. To specify a different primary input channel, use the --iterate-primary-input-channel parameter. An integer value is expected, where the first channel is specified with the value 0, the second channel is specified with the value 1, and so on.
For further information about defining batch units, see the CLC Genomics Workbench manual.
Launching workflows containing Collect and Distribute elements
The most common situation is to have a Collect and Distribute element with a single input channel and a single output channel, as is the case in the example in figure 8.3. With this design, the results from all the batch units in the upstream iteration block are collected and passed on together as input to the connected downstream step(s). Such Collect and Distribute elements do not require any parameters to be defined on the command line.
Where the Collect and Distribute element has more than one output channel, the parameters below must be specified. Parameter names start with the workflow element name, which in this case was the default name, Collect and Distribute.
--collect-and-distribute-group-by-metadata-columnProvide the name of the metadata column to use to group the data, where each group is sent individually as input to the connected downstream analysis step(s). The values in this column are mapped to the relevant Collect and Distribute output channel using the parameter below.--collect-and-distribute-output-mappingDefine the Collect and Distribute output channel that data should flow through by mapping each value in the relevant metadata column, specified using the parameter above, to an output channel. The format required is'<sample-information>=<output channel name>'. For example, if samples with a metadata value "Treated" should flow through an output channel called "Type 1", the value would be'Treated=Type 1'. One parameter-value pair is needed for each mapping.
Template workflow example using Iterate and Collect and Distribute elements
The RNA-Seq and Differential Gene Expression Analysis template workflow, distributed with the CLC Genomics Workbench, provides an example of using Iterate and Collect and Distribute elements. It is described in detail in the CLC Genomics Workbench manual.
