Extracting alignments
If you wish to compare the sequences of two or more sequences types in a multilocus sequence analysis, you can easily extract concatenated alignments from the profile table. To extract an alignment displaying the sequences of one or more loci:
Select the sequence types you wish to align | right-click | Extract Alignment | Sequence Types or Alleles
The two options in the alignment right-click menu provide two ways of composing the alignment (figure 3.12):
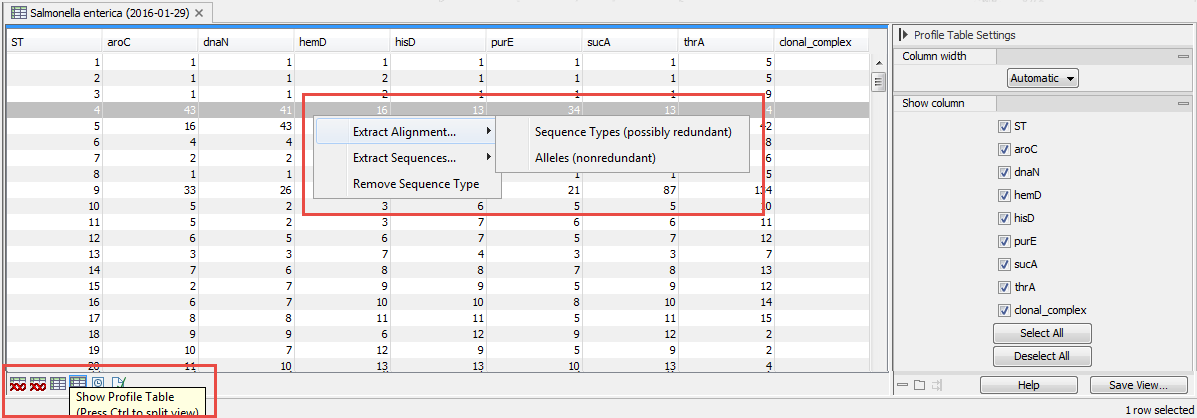
Figure 3.12: Extract alignment from a profile table.
- Sequence Types (possibly redundant). An alignment is made with alleles from all the selected sequence types, even if the same allele occurs more than once.
- Alleles (non-redundant). Redundant alleles are excluded from the alignment where possible. If you e.g. select three rows in the table and two of the sequence types have the same allele sequence in all the selected loci, the alignment will only include two sequences (the first option would have included all three sequences in the alignment).
Selecting either option will display a dialog similar to the one shown in figure 3.13 displaying the loci in the scheme.
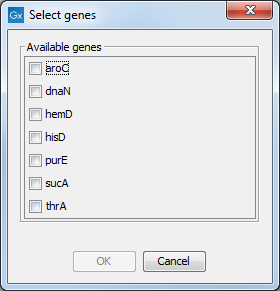
Figure 3.13: Selecting genes for alignment.
Before calculating the alignment, the allele sequences for each sequence type will be joined into a "supergene" sequence which is used for the alignment. The checkboxes shown in figure 3.13 let you specify which of the loci that should be used in the joined sequence. Selecting only one locus will create a "non-supergene" alignment with allele sequences for only this particular locus.
When you click OK, a dialog with alignment parameters will be shown. To learn more about these parameters, click the dialog's Help button.
