Assemble to an existing isolate
CLC MLST Module makes it easy to perform sequence typing in several iterations. If you e.g. find errors in the sequencing data of some of the loci, you can create a preliminary version of the isolate with only sequence data for some of the loci. Then you can re-sequence the missing loci and update the isolate with new sequences.
This is done using the Assemble to Existing Isolate tool, available from:
Tools | Multilocus Sequence
Typing (![]() )| Assemble to Existing Isolate (
)| Assemble to Existing Isolate (![]() )
)
As input for this tool, select exactly one isolate and at least one nucleotide sequence. It is possible to select a nucleotide sequence list (see figure 4.6) or multiple nucleotide sequences.
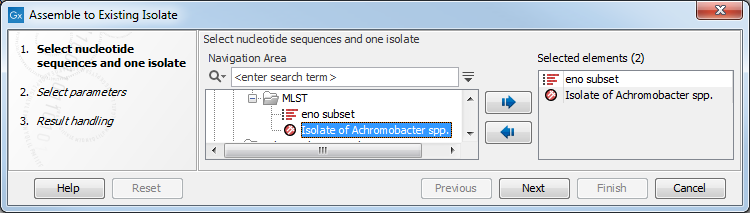
Figure 4.6: Assembling an isolate.
The rest of the dialogs are identical to the dialogs displayed when creating an isolate de novo (see Assemble and create isolate).
