Single Cell TCR-Seq Analysis
Single Cell TCR-Seq Analysis can be found in the Toolbox here:
Immune Repertoire (![]() ) | Single Cell TCR-Seq Analysis (
) | Single Cell TCR-Seq Analysis (![]() )
)
The tool takes as input one or more lists of reads that have been annotated using Annotate Reads with Cell and UMI. It outputs a Cell Clonotypes (![]() ) element, and optionally a report.
) element, and optionally a report.
|
Note: Different runs can result in slightly different results. This is caused by multi-threading of the program combined with the use of probabilistic data structures. The overall content of the Cell Clonotypes should not be markedly different.
Barcode whitelists: In some protocols, the set of valid barcodes is known in advance, and available as a barcode whitelist. In CLC Single Cell Analysis Module, it is not possible to directly use such a list. Instead, the Filter Cell Clonotypes can be used for filtering the Cell Clonotypes output such that only barcodes that are identified as cells are retained, such as those identified as cells in matched scRNA-Seq data. Additionally, the Filter Cell Clonotypes can be used for retaining only the desired types of clonotypes, for example only those that are productive. See Filter Cell Clonotypes for details. |
It is important to provide all the data for a sample to the tool at the same time. For instance, if one sample was sequenced on 4 lanes of an Illumina sequencer, then all 4 lanes should be supplied together. This allows reads originating from the same cell but coming from different lanes to be analyzed jointly and leads to a more accurate clonotype identification.
The following options can be adjusted (figure 10.1):
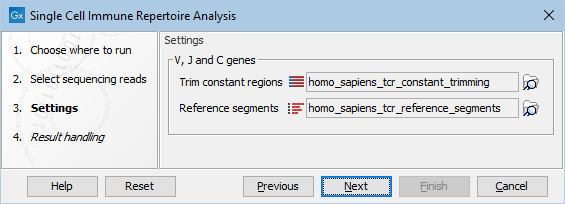
Figure 10.1: The options in the dialog of the Single Cell TCR-Seq Analysis tool. Human reference data downloaded from the Reference Data Manager has been selected.
- Trim constant regions. The C (constant) gene segments, provided as a Trim Adapter List. These regions are trimmed prior to the clonotype identification.
- Reference segments. The V (variable) and J (joining) gene segments. These are used during during clonotype identification.
Both elements are reference data elements and need to be downloaded from the Reference Data Manager (see The Reference Data Manager).
Subsections
- The report output from Single Cell TCR-Seq Analysis
- The Cell Clonotypes element
- The clonotype identification algorithm
