Importing matrices
As with any workflow, the Expression Matrix or Peak Count Matrix or both can be imported with on-the-fly imports. There are a number of specific things to be aware of here, though.As already mentioned, the Expression Matrix and Peak Count Matrix must be for cells originating from the same sample. This will be the case if they have been exported, the barcode format was set to include the sample and they were referring to the same sample prior to exporting (see Export Peak Count Matrix).
If the barcodes don't include the sample it is instead taken from the name of the file. The files must then be named so they match the sample, if necessary moving expression and peak files into separate folders as illustrated in figure 16.6.
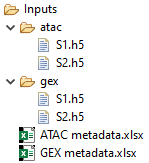
Figure 16.6: Naming files to match sample
Another thing to be aware of is that if supplying nearby genes and/or transcription factors in separate files the HDF5 importer is limited to taking just one file as shown in figure 16.7 and the same file is then used for all imported files (samples). This means that in practice the HDF5 importer is not suitable for running the workflow with multiple samples if nearby genes and/or transcription factors are explicitly supplied.
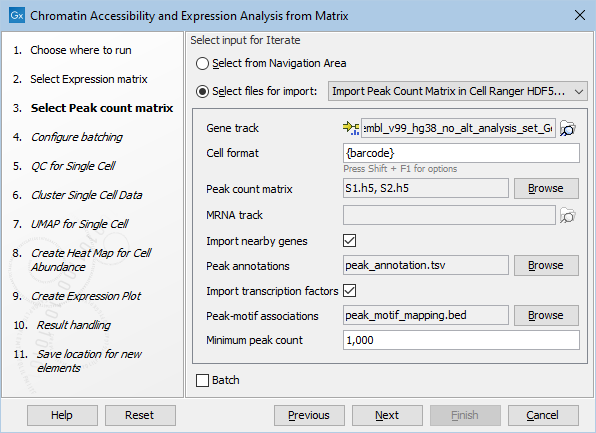
Figure 16.7: Specifying nearby genes and transcription factors for HDF5
Instead, the archive MEX format can be used as shown in figure 16.8. It allows bundling separate nearby genes and/or transcription factors in each archive. For more information, see Import Peak Count Matrix.
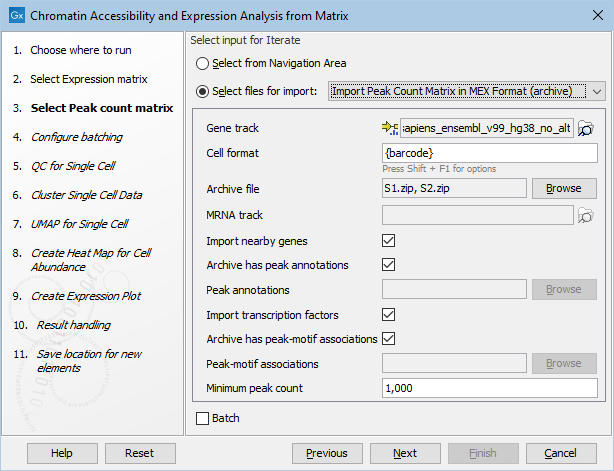
Figure 16.8: Specifying nearby genes and transcription factors for MEX
