Configuring the batch units for Chromatin Accessibility and Expression Analysis from Reads
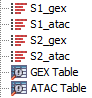
Figure 15.10: ATAC and GEX reads for two samples along with metadata
Assume as an example that the input consists of ATAC and GEX reads for two samples as shown in figure 15.10. The reads must be linked by metadata tables. That is, there must be one metadata table with all ATAC reads and one with all GEX reads and the two must have a common column linking them (e.g., Sample).
The batching can be configured as shown in figure 15.11. In this example:
- The batching is based on the column "Sample" in the metadata table for the ATAC reads. There will be one iteration per distinct value, which typically means per row.
- The ATAC and GEX reads are matched by the same "Sample" column. It must be present in both metadata tables. The matching column does not need to be the same as the batching column, but it typically is.
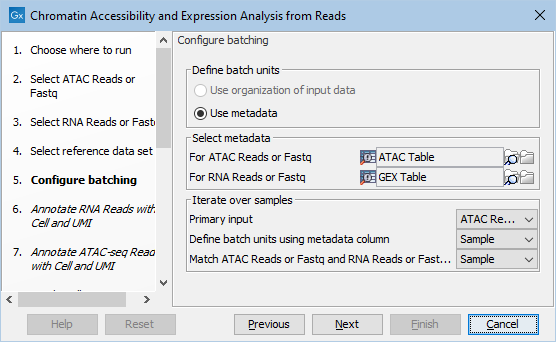
Figure 15.11: Configuring batching of coupled ATAC and GEX reads
There will be two iterations, one for the ATAC and GEX reads for sample S1 and one for S2 as shown in figure 15.12.
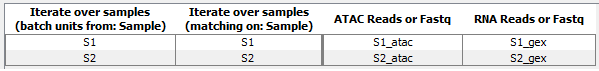
Figure 15.12: Batch overview for coupled ATAC and GEX reads
It is also possible to use the same metadata table for ATAC and GEX reads. Then it must be selected twice in the batch configuration step.
