Open reading frame parameters
This opens the dialog displayed in figure 16.8:
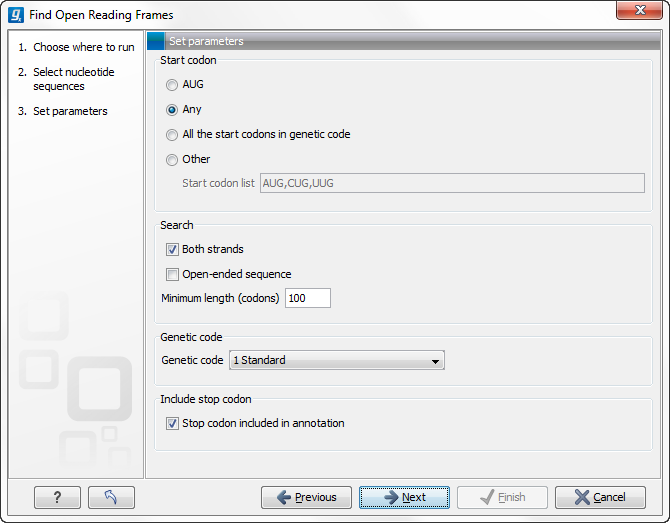
Figure 16.8: Create Reading Frame dialog.
The adjustable parameters for the search are:
- Start codon:
- AUG. Most commonly used start codon.
- Any. Find all open reading frames of specified length. Any combination of three bases that is not a stop-codon is interpreted as a start codon, and translated according to the specified genetic code.
- All start codons in genetic code.
- Other. Here you can specify a number of start codons separated by commas.
- Both strands. Finds reading frames on both strands.
- Open-ended Sequence. Allows the ORF to start or end outside the sequence. If the sequence studied is a part of a larger sequence, it may be advantageous to allow the ORF to start or end outside the sequence.
- Genetic code translation table.
- Include stop codon in result The ORFs will be shown as annotations which can include the stop codon if this option is checked. The translation tables used are listed in here.
- Minimum Length. Specifies the minimum length for the ORFs to be found. The length is specified as number of codons.
Using open reading frames for gene finding is a fairly simple approach which is likely to predict genes which are not real. Setting a relatively high minimum length of the ORFs will reduce the number of false positive predictions, but at the same time short genes may be missed (see figure 16.9).
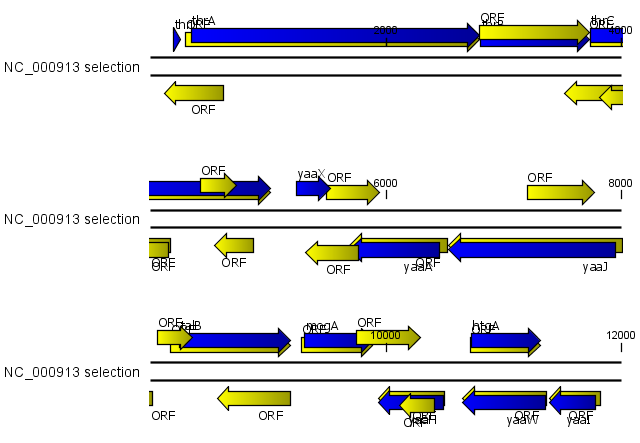
Figure 16.9: The first 12,000 positions of the E. coli sequence NC_000913 downloaded from GenBank. The blue (dark) annotations are the genes while the yellow (brighter) annotations are the ORFs with a length of at least 100 amino acids. On the positive strand around position 11,000, a gene starts before the ORF. This is due to the use of the standard genetic code rather than the bacterial code. This particular gene starts with CTG, which is a start codon in bacteria. Two short genes are entirely missing, while a handful of open reading frames do not correspond to any of the annotated genes.
Click Next if you wish to adjust how to handle the results. If not, click Finish.
Finding open reading frames is often a good first step in annotating sequences such as cloning vectors or bacterial genomes. For eukaryotic genes, ORF determination may not always be very helpful since the intron/exon structure is not part of the algorithm.
