The Detect QIAseq MSI Status ready-to-use workflow
The Detect QIAseq MSI Status workflow has been designed to support the DHS-8800Z QIAseq Targeted DNA panels sequenced using Illumina technology. This particular workflow includes only the Detect MSI Status tool, and it was created in order to allow easy execution directly from the Analyze QIAseq Panels guide. The Detect MSI Status tool measures the statistical variation of the length distribution of each microsatellite locus and decides for each locus if it is stable or not by comparing the statistical variation of the test sample with the normal samples' baseline. If the proportion of unstable microsatellite loci is higher than a predefined threshold, then the sample is considered unstable.
To learn more about the workflow, read about the tool in Detect MSI status.
To run the workflow outside of the Analyze QIAseq Panels guide, go to:
Ready-to-Use Workflows | QIAseq Panel Analysis (![]() ) | QIAseq Analysis Workflows (
) | QIAseq Analysis Workflows (![]() ) | Detect QIAseq MSI Status (
) | Detect QIAseq MSI Status (![]() )
)
The Detect QIAseq MSI Status workflow will take as input a read mapping previously generated by running the Identify TMB Status workflow.
Next, you need to select the appropriate Reference Data Set (figure 5.19).
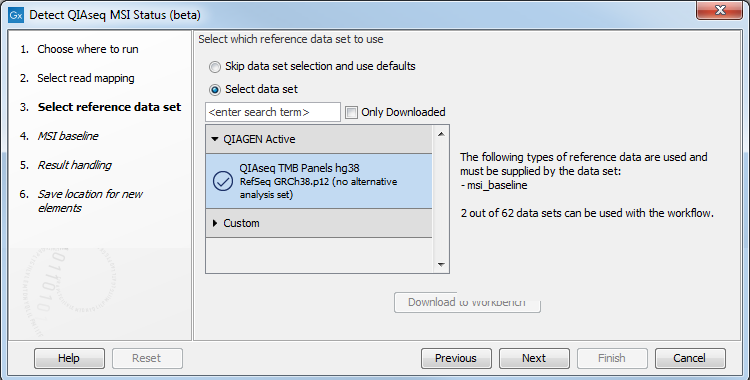
Figure 5.19: Select the appropriate Reference Data Set.
The Reference Data Manager includes the QIAseq TMB Panels hg38 (no alternative analysis set) set. This data set includes two ready-to-use baseline tracks: one is based on a 27 microsatellite loci track containing all the microsatellite loci covered by the primers in the Human TMB and MSI Panel (DHS-8800Z). This baseline was created using 30 MSS samples that were mapped to the hg38 (no alternative analysis set) reference sequence and processed with the Generate MSI Baseline tool using the default parameters. The other is a subset of the first containing 9 loci. These loci were identified utilising lung FFPE MSS and MSI samples and were found to perform consistently well during benchmarking (figure 5.20). All 9 loci are mono nucleotide homopolymers.
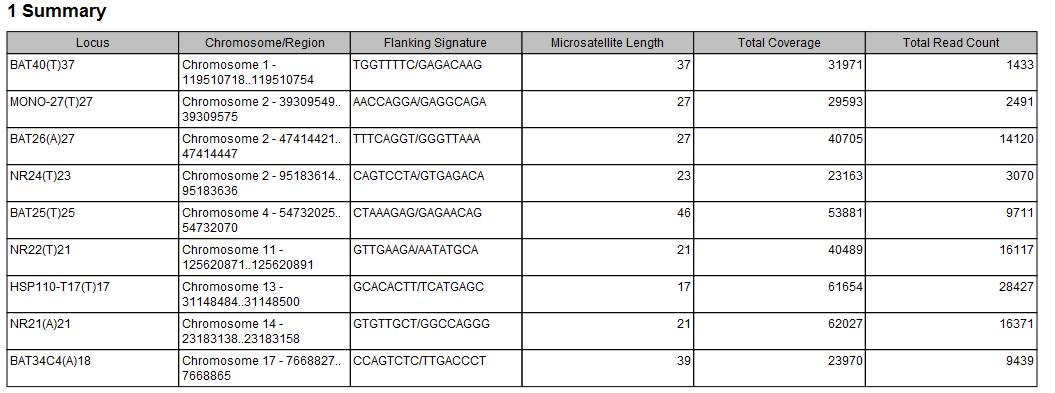
Figure 5.20: Report generated when creating the 9 loci MSI baseline.
The 27 loci baseline track should not be used for detecting MSI status. This baseline was included in the Reference Data Set to allow the selection of a subset of loci specific to a particular cancer type. To generate a cancer-specific baseline track, open a Track List of the 27 loci baseline track along with your samples read mappings to investigate quality of the loci of interest. Then select the loci that should be included in the baseline (in the table view of the 27 loci baseline track) and click on the Create Track from Selection button at the bottom of the table. Save the newly generated baseline in the Navigation Area, and create a custom Reference Data set by replacing the msi_loci and msi_baseline element with the track you just generated.
Also note that the samples were sequenced using NextSeq (Illumina) and will work best against samples sequenced on this sequencer. Similarly, when using Ion Torrent reads we recommend to create a custom baseline from at least 15 normal samples compatible with the samples of interest, i.e., samples from the same population and sequenced on same equipment and in the same lab to minimize sample preparation biases.
Subsections
