Structural Variant Caller output
The tool has the following output options, as shown in figure 9.2:
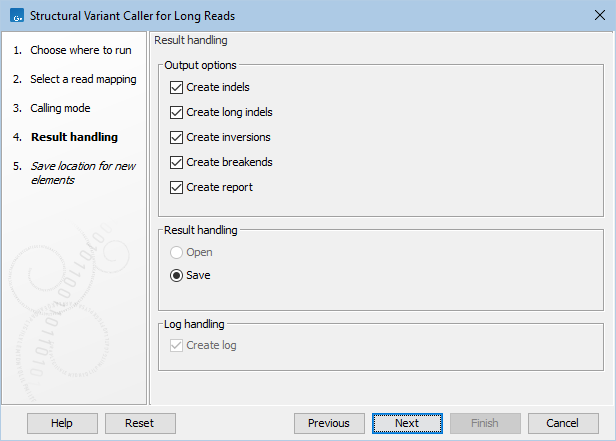
Figure 9.2: The output options for Structural Variant Caller for Long Reads.
- Indels (Indels) (
 ). A variant track with indels (deletions and insertions - including tandem duplications) that have lengths up to 100,000 bp.
). A variant track with indels (deletions and insertions - including tandem duplications) that have lengths up to 100,000 bp.
- Long indels (Indels long) (
 ). An annotation track with long indels (those with lengths larger than 100,000 bp). The reason for putting indels larger than 100,000 bp in a separate annotation track, is that these variants have either long allele or reference entries in the variant track, which make them challenging to work with in the track viewer.
). An annotation track with long indels (those with lengths larger than 100,000 bp). The reason for putting indels larger than 100,000 bp in a separate annotation track, is that these variants have either long allele or reference entries in the variant track, which make them challenging to work with in the track viewer.
- Inversions (Inv) (
 ). An annotation track with inversions.
). An annotation track with inversions.
- Breakends (Breakends) (
 ). An annotation track with a row for each breakend in a translocation.
). An annotation track with a row for each breakend in a translocation.
- Report (
 ). A report giving an overview over analyzed references and found structural variants.
). A report giving an overview over analyzed references and found structural variants.
All outputs (other than the Report) can be exported together to a single VCF file, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Export_in_VCF_format.html. The VCF-exportable outputs contain the following annotations:
- Chromosome. The chromosome on which the variant is located.
- Region. The location of the variant.
- Zygosity. The zygosity of the variant called, as determined by the variant detection tool. This will be either 'Homozygous', where there is only one variant called at that position or 'Heterozygous' where more than one variant was called at that position.
- Count. The estimated count for the structural variant. In the case of insertions longer than 2500 bp, this is increased by an ad hoc procedure to account for reads that are not observed because they align within the insertion. If the coverage cannot be determined then this is reported as '0'. It is therefore advised to perform any desired filtering of variants on the 'Raw count' annotation rather than this annotation.
- Coverage. The estimated coverage for the structural variant. This is determined in different ways for different structural variant types. For insertions, this is the coverage at the location of the insertion; for duplications it is the average of the coverage at the two ends of the duplication; for inversions it is the average at positions upstream and downstream of the inversion; for deletions and breakends it is the average of the coverage at the start, end, and center of the variant. When taking averages, locations with no reads are ignored. In rare cases the coverage cannot be determined, because none of the locations used for calculating it have any reads. In these cases, the coverage will be reported as '0'.
- Frequency. The count divided by the coverage, reported as a percentage.
- Average quality. The average mapping quality of the alignments supporting the variant. The mapping quality is reported on the Phred scale, and describes the probability that the alignment is incorrect. The estimates are made by minimap2. A value of 30 means a 1 in 1000 chance of incorrect alignment. Very low average qualities are not seen, because the algorithm filters alignments with low mapping quality. Note that other variant calling tools, such as the Low Frequency Variant Detection tool, use this annotation to report the average quality scores of reads on the Phred scale rather than the average quality of alignments on the Phred scale.
- Stdev position. An estimate of the standard deviation of the position of the variant.
Additional annotations present on more than one output are:
- Length. The length of the variant. For deletions, it is the length of the deleted sequence, and for insertions and duplications it is the length of the inserted sequence. For inversions, it is the length of the inverted region.
- Allele. The inserted sequence. This is only reported for insertions and duplications.
- Forward read count. For inversions, the number of countable reads supporting the 5' side of the inversion on the reference. For other types of variant, the number of reads supporting the allele and mapping in the forward direction. The 'countable' reads are those that are used by the variant detection tool when calling the variant.
- Reverse read count. For inversions, the number of countable reads supporting the 3' side of the inversion on the reference. For other types of variant, the number of reads supporting the allele and mapping in the reverse direction. The 'countable' reads are those that are used by the variant detection tool when calling the variant.
- Raw count. The number of countable reads supporting the allele. The 'countable' reads are those that are used by the variant detection tool when calling the variant.
- Detailed type. This is only present for insertions and is set to "Insertion" if the variant describes a novel insertion, and "Duplication" if the insertion instead describes a duplication.
- Repeat unit size. Duplications are sometimes detected as insertions within a read. In these cases, their reported "Length" is estimated from these insertions. Because the reference sequence may be duplicated more than once, this can be larger than the duplicated region on the reference. This is an estimate of the size of one copy on the reference. It is equal to the "Length" if it cannot be estimated. In rare circumstances, the "Repeat unit size" may be estimated to exceed the "Length". This is usually caused when a repeat is inserted between two existing repeats of the same kind.
- Stdev length. An estimate of the standard deviation of the length of the variant.
Indels variant track
The indels track uses many of the standard variant annotations, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Variant_tracks.html.
Long indels variant track
This track contains insertions and deletions larger than 100,000 bp. It is often enriched for false positive calls. This is because deletions and duplications are called when a read maps to two disjoint locations. If these two locations are at either end of a chromosome, then a near-whole chromosome deletion (or duplication) will be called. In many cases, it is more likely that the read maps to two places because an insertion is present that shares homology with one of the locations.
It is sometimes possible to detect false positives. For example, if the sample is germline, and other structural variants are called within a long homozygous deletion, then it is likely that the long homozygous deletion is a false positive.
Inversions track
This track is often enriched for false positive calls. This is because an inversion may be called when a read maps to two disjoint locations on the same chromosome and in different orientations. If these two locations are at either end of a chromosome, then a near-whole chromosome inversion will be called. In many cases, it is more likely that the read maps to two places because an insertion is present that shares homology with one of the locations.
When coverage is high, it is often possible to detect false positives by requiring that there is support for both sides of the inversion. Reads supporting the 5' side of the inversion on the reference are counted as "forward" reads, and reads supporting the 3' side of the inversion on the reference are counted as "reverse" reads. Each variant reports these in "Forward read count" and "Reverse read count" annotations respectively.
Another class of false positives are inversions that start or end at the same location as an insertion. This is sometimes a signature of an inverted repeat.
Breakend track
The breakend track can be used to look for translocations and other complex rearrangements that involve more than one chromosome. The definition of a breakend that we use here closely follows that from the VCF specification. Please refer to Section 5.4 "Specifying complex rearrangements with breakends" of https://samtools.github.io/hts-specs/VCFv4.4.pdf. Specifically we support the cases shown in figures 1, 4, 5, and 7 of that section.
Annotations that are only present on the breakends output are:
- Filter. This is always PASS. It is necessary for VCF export.
- Breakpoint type. 5' if the breakend is on an earlier chromosome than its mate, 3' if it is on a later chromosome.
- Type. Either "donor" or "acceptor". Reads mapping to a "donor" breakend are mapped to the left of the breakend, disappear at the breakend location, and reappear on another chromosome. Reads mapping to an "acceptor" breakend appear at the breakend location and continue to the right of the location.
- Fusion crossing reads. The number of distinct reads that support the breakend. Note that this is not necessarily the number of reads that support the fusion, because the same breakend may take part in more than one fusion.
- Fusion number. Two breakends that share a fusion number describe one fusion. The read aligns up to one breakend on one chromosome, and then continues aligning at the other breakend on a different chromosome. If a breakend is involved in more than one fusion, it will appear more than once in the track, with a different fusion number for each fusion.
A simple reciprocal translocation involves 4 breakends: an acceptor and donor on each of the two chromosomes involved in the translocation. The 4 breakends will have different combinations of Name and Type, and two different Fusion numbers.
The easiest way to find translocations is to:
- Open the table view of the Breakend track and sort by Fusion number by clicking the column header.
- Look for two nearby fusion pairs in the sorted table with the same Chromosome and similar Region.
- Verify that each fusion pair consists of one donor and and one acceptor type, and that each chromosome contains one donor and one acceptor type.
Note that the Region for a breakend is sometimes on the plus strand (e.g. 123456^123457) and sometimes on the minus strand (e.g. complement(123456^1234567)). There is no significance to the reported strand - it is used by the VCF exporter.
