Typing of samples among multiple species
The Type Among Multiple Species workflow is designed for typing a sample among multiple predefined species (figure 8.19).
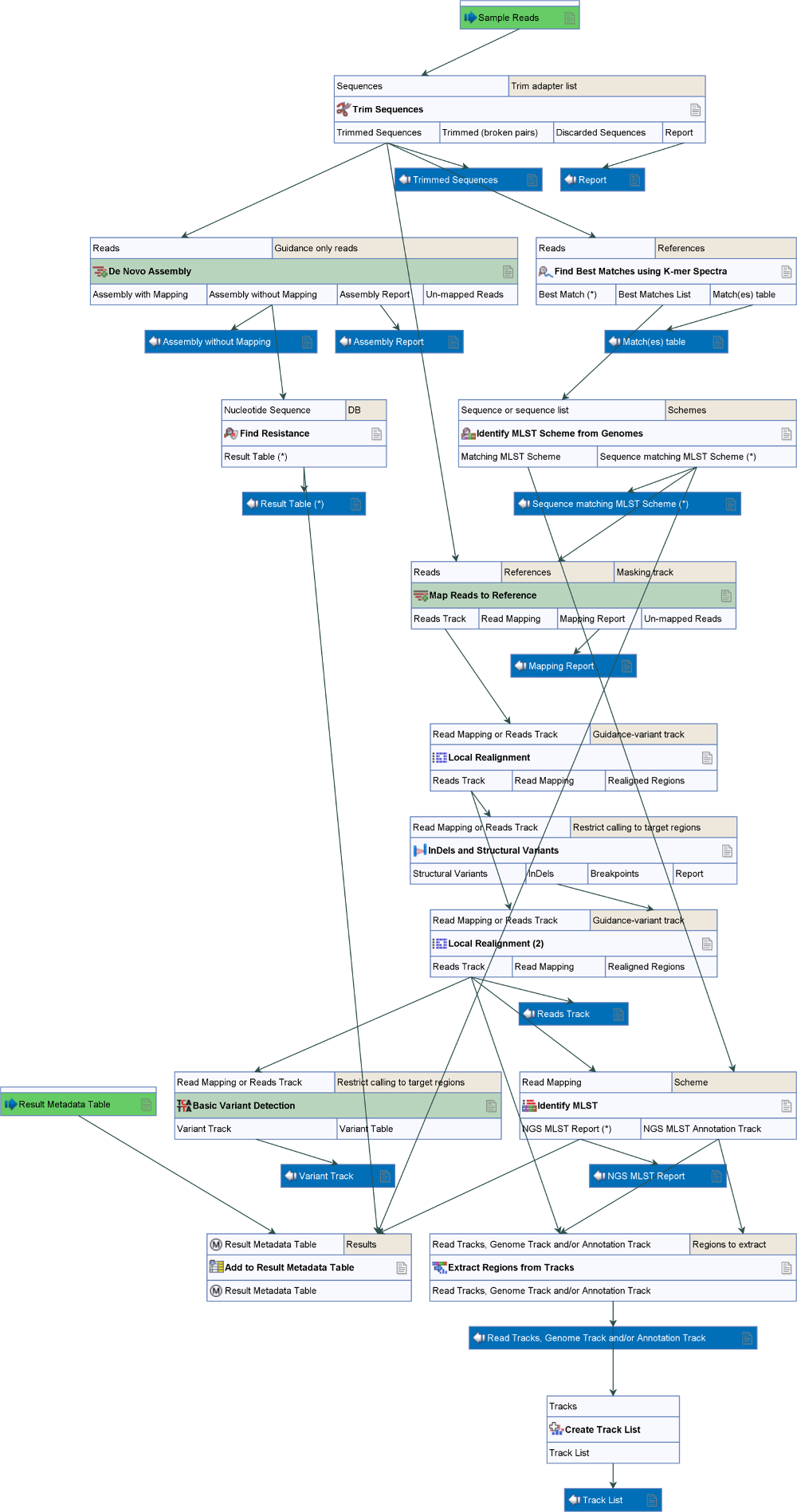
Figure 8.19: Overview of the template Type Among Multiple Species workflow.
Similar to the Type a Known Species workflow, it allows identification of the closest matching reference species among the user specified reference list(s), but in this case, the list(s) may represent multiple species. The workflow identifies the associated MLST scheme and type, determines variants found when mapping the sample data against the identified best matching reference, and finds occurring resistance genes if they match genes within the user specified resistance database.
The workflow also automatically associates the analysis results to the user specified Result Metadata Table. For details about searching and quick filtering among the sample metadata and generated analysis result data, see section 9.4.1.
Subsections
- Preliminary steps to run the "Type Multiple Species" workflow
- How to run the "Type Among Multiple Species" workflow for a single sample
- Example of results obtained using the Type Among Multiple Species workflow
- How to run the "Type Among Multiple Species" workflow on a batch of samples:
