Estimate Alpha and Beta Diversities workflow
The Estimate Alpha and Beta Diversities workflow consists of 5 tools and requires only the OTU table as input file (figure 6.2). Remember to add metadata to your output table before starting the workflow.
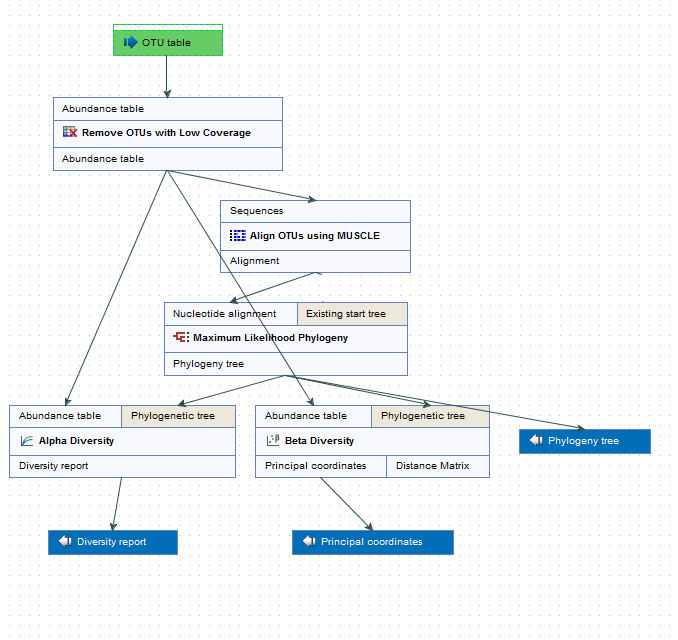
Figure 6.2: Layout of the Alpha and Beta Diversities workflow.
The first tool of the workflow is the Filter OTUs Based on the Number of Reads. The output is a reduced abundance table that will be used as input for 3 other tools:
- Align OTUs with MUSCLE, a tool that will produce an alignment used to reconstruct a Maximum Likelihood Phylogeny (see http://www.clcsupport.com/clcgenomicsworkbench/current/index.php?manual=Maximum_Likelihood_Phylogeny.html), which will in turn output a phylogenetic tree also used as input in the following 2 tools.
- Alpha diversity tool
- Beta diversity tool
Running this workflow will therefore give 3 outputs: a phylogenetic tree of the OTUs, a diversity report for the alpha diversity and a PCoA for the beta diversity.
