OTU clustering parameters
After having selected the sequences you would like to cluster, the wizard offers to set some general parameters (see figure 3.3).
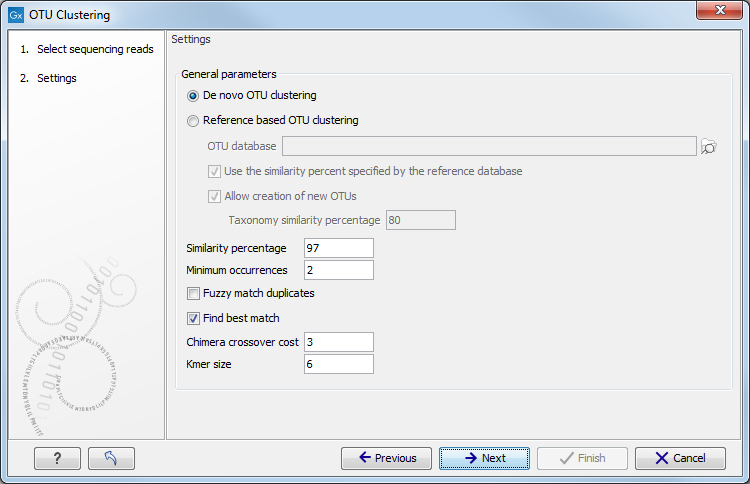
Figure 3.3:
Settings for the OTU clustering tool.
You can choose to perform a De novo OTU clustering, or you can perform a Reference based OTU clustering.
The following parameters can then be set:
- OTU database Specify here the reference database to be used for Reference based OTU clustering. Reference databases can be created by the Download OTU Reference Database or the Format Reference Database tools.
- Use the similarity percent specified by the reference database Allows to use the same similarity percent value (see below) that was used when creating the reference database. This parameter is available only when performing a reference based OTU clustering. Selecting this parameter will disable the similarity percent parameter.
- Allow creation of new OTUs Allows sequences which are not already represented at the given similarity distance in the database to form a new cluster, and a new centroid is chosen. This parameter can be set only when performing a Reference based OTU clustering. Disallowing the creation of new OTUs is also known as closed reference OTU picking.
- Taxonomy similarity percentage Specifies the similarity percentage to be used when annotating new OTUs. This parameter is available only when Allow creation of new OTUs is selected.
- Similarity percentage: Specifies the required percentage of identity between a read and the centroid of an OTU for the read to join the OTU cluster.
- Minimum occurrences: Specifies the minimum number of duplicates for specific read-data before it will be included in further analyses. For instance, if set to 2, at least two reads with the same exact nucleotides needs to exist in the input for the data to propagate to further analysis. Other data will be thrown away. This can for instance be used to filter out singletons. Note that matches does not need to be exact when the Fuzzy match duplicates option is used.
- Fuzzy match duplicates: Specifies how duplicates are defined. If the option is not selected two reads are only duplicates if they are exactly equal. If the option is selected, two reads are duplicates if they are almost equal, i.e. all differences are SNVs and there are not too many of them (
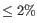 ). This pseudo-merging is done by lexicographically sorting the input and looking in the neighborhood of the read being processed. The reads are processed from most abundant (in a completely equivalent sense) to the least. In this way two singletons can for instance be pseudo-merged together and be included for further study despite the Minimum occurrences option having specified 2. Upon further analysis a group can be split into several OTUs if not all members are within the specified threshold from the "OTU-leader".
). This pseudo-merging is done by lexicographically sorting the input and looking in the neighborhood of the read being processed. The reads are processed from most abundant (in a completely equivalent sense) to the least. In this way two singletons can for instance be pseudo-merged together and be included for further study despite the Minimum occurrences option having specified 2. Upon further analysis a group can be split into several OTUs if not all members are within the specified threshold from the "OTU-leader".
- Find best match: If the option is not selected, the read becomes a member of the first OTU-database entry found within the specified threshold. If the option is selected all database entries are tested and the read becomes a member of the best matching result. Note that "first" and "all" are relative terms in this case as kmer-searches are used to speed up the process. "All" only includes the database entries that the kmer search deems close enough, i.e., database entries that cannot be within the specified threshold will be filtered out at this step. "First" is the first matching entry as returned by the kmer-search which will sort by the number of kmer-matches.
- Chimera crossover cost: The cost of doing a chimeric crossover, i.e. the higher the cost the less likely it is that a read is marked as chimeric.
- Kmer size: The size of the kmer to use in regards to the kmer usage in finding the best match.
Chimera detection is performed by kmer searches as follows:
- All database entries and the read being processed are split into 4 equally sized portions with an additional 3 half-way-shifted to cover the merge-points. This results in 7 different kmer-search-options. Each of these is queried for matches within some threshold, and only the results are processed further. A database entry has to fit well in at least one of these 7 portions for the database-entry to be relevant.
- Given the 7 sets of database entries some entries may just be present in one of the sets. For these entries, some may be duplicates in the region they represent. The duplicates are filtered out and only an arbitrary representative of the duplicates is kept.
