Tracks
A track is the fundamental building block for NGS analysis in the CLC Genomics Workbench. The idea behind tracks is to provide a unified framework for the visualization, comparison and analysis of genome-scale studies such as whole-genome sequencing or exome resequencing projects and a variety of different -Seq data (i.e. ChIP-Seq, DNAse-Seq).24.1
In tracks, all information is tied to genomic positions. A central coordinate-system is provided by a reference genome, which allows that different datasets can be seen and analyzed together. Different kinds of tracks exist: a reference genome sequence (![]() ), a set of genes (
), a set of genes (![]() ), a coverage graph (
), a coverage graph (![]() ), a read mapping (
), a read mapping (![]() ) or variants from variant calling (
) or variants from variant calling (![]() ). This chapter explains how to visualize tracks, how to retrieve reference data and finally how to perform generic comparisons between tracks.
). This chapter explains how to visualize tracks, how to retrieve reference data and finally how to perform generic comparisons between tracks.
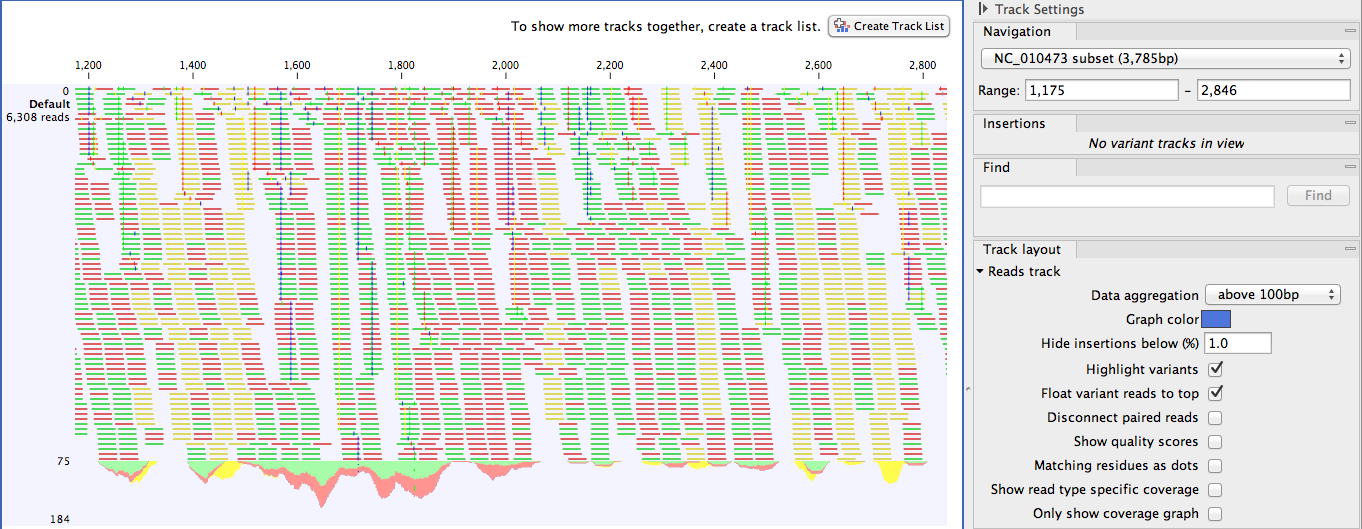
Figure 24.1: A single mapping read-track opened, displaying reads and coverage. On the top right, the button for creating a Track List is visible. On the right is the SidePanel.
For comparison tools specific to resequencing and variants, please see Resequencing.
Footnotes
- ... DNAse-Seq).24.1
- The track concept was first introduced with the Genomics Gateway plugin in 2011 and made an integral part of the CLC Genomics Workbench 5.5 release.
Subsections
- Track lists
- Retrieving reference data tracks
- Merging tracks
- Converting data to tracks and back
- Annotate and filter tracks
- Creating graph tracks
