Local realignment
The goal of the local realignment tool is to improve on the alignments of the reads in an existing read mapping. The local realignment algorithm works by exploiting the information available in the alignments of other reads when it is attempting to re-align any given read. Most mappers do not use cross-read information as it would be computationally prohibitive to do within the mapping algorithm. However, once the reads have been mapped, local realignment procedures can exploit this information.
Realignment will typically occur in areas around insertions and deletions in the sample reads relative to the reference. In such regions we wish to see our reads mapped with one end of the read on one side of the indel and the rest mapped on the other side. However, the mapper that originally mapped the reads to the reference does not have information about the existence of an indel to use when mapping a given read. Thus, reads that are mapped to such regions, but that only have a short part of the read representing the region on one side of the indel, will typically not be mapped properly across the indel, but instead be mapped with this end unaligned, or into the indel region with many mismatches. The Local Realignment tool can use information from the other reads mapping to a region containing an indel, including reads that are located more centered across the indel and thus have been mapped with ends on either side of the indel. As a result an alternative mapping, as good as or better than the original, can be generated.
Local realignment will typically have an effect on any read mapping, whether the reads were mapped using a local or global alignment algorithm (i.e. with the Global alignment option of the mapping tool unchecked (the default) or checked, respectively). An example of the effect of using the Local Realignment tool on a read mapping made using the the local alignment algorithm is shown in figure 25.26. An example in the case of a mapping made using the global alignment algorithm is shown in figure 25.27.

Figure 25.26: Local realignment of a read mapoping produced with the 'local' option. [A] The alignments of the first, second, and fifth read in this read mapping do not support the four-nucleotide insertion supported by the remaining reads. A variant caller might be tempted to call a heterozygous insertion of four nucleotides in one allele and heterozygous replacement of four nucleotides in a second allele. [B] After applying local realignment, the first, second, and fifth read consistently support the four-nucleotide insertion.
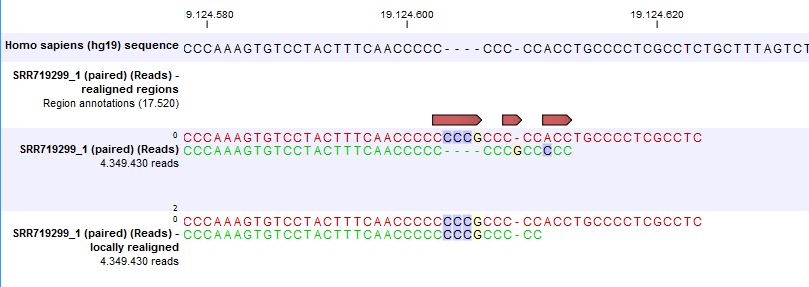
Figure 25.27: Local realignment of a read mapoping produced with the 'global' option. Before realignment the green read was mapped with two mismatches. After realignment it is mapped with the inserted 'CCCG' sequence (seen in the alignment of the red read) and no mismatches.
Subsections
- Method
- Realignment of unaligned ends
- Guided Realignment
- Multi-pass local realignment
- Known Limitations
- Computational Requirements
- How to run the Local Realignment tool
