An example using Illumina barcoded sequences
The data set in this example can be found at the Short Read Archive at NCBI: http://www.ncbi.nlm.nih.gov/sra/SRX014012.
It can be downloaded directly in fastq format via the URL http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?cmd=dload&run_list=SRR030730&format=fastq. The file you download can be imported directly into the Workbench.
The barcoding was done using the following tags at the beginning of each read: CCT, AAT, GGT, CGT (see supplementary material of [Cronn et al., 2008] at http://nar.oxfordjournals.org/cgi/data/gkn502/DC1/1).
The settings in the dialog should thus be as shown in figure 19.29.
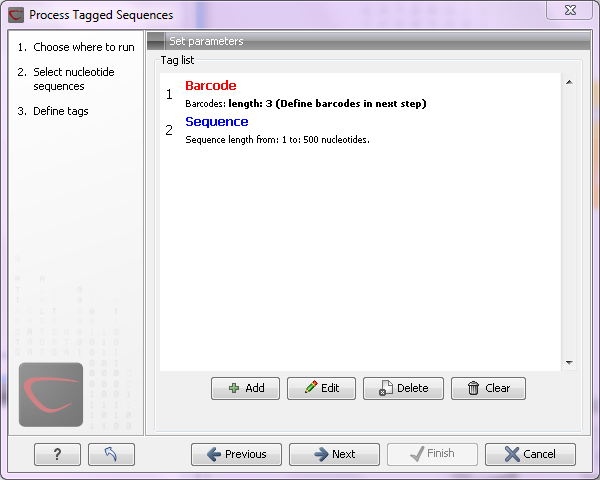
Figure 19.29: Setting the barcode length at three
Click Next to specify the bar codes as shown in figure 19.30 (use the Add button).
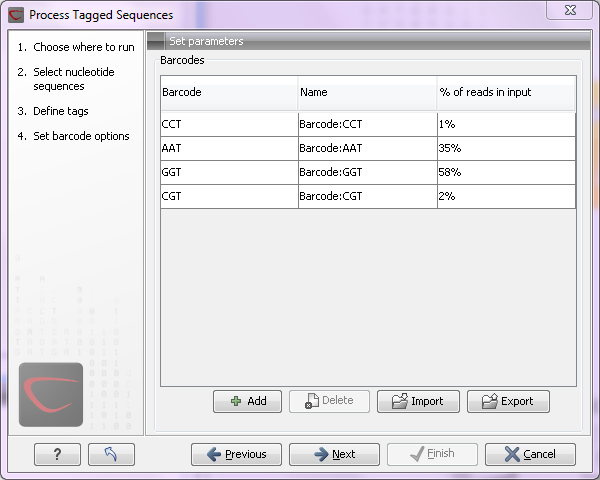
Figure 19.30: A preview of the result
With this data set we got the four groups as expected (shown in figure 19.31). The Not grouped list contains 445,560 reads that will have to be discarded since they do not have any of the barcodes.
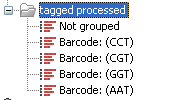
Figure 19.31: The result is one sequence list per barcode and a list with the remainders
