View Annotations in a table
Annotations can also be viewed in a table:
Select a sequence in the Navigation Area and right-click on the file name | Hold the mouse over "Show" to enable a list of options | Annotation Table (![]() )
)
or If the sequence is already open | Click Show
Annotation Table (![]() ) at the lower left part of
the view
) at the lower left part of
the view
This will open a view similar to the one in figure 9.13).
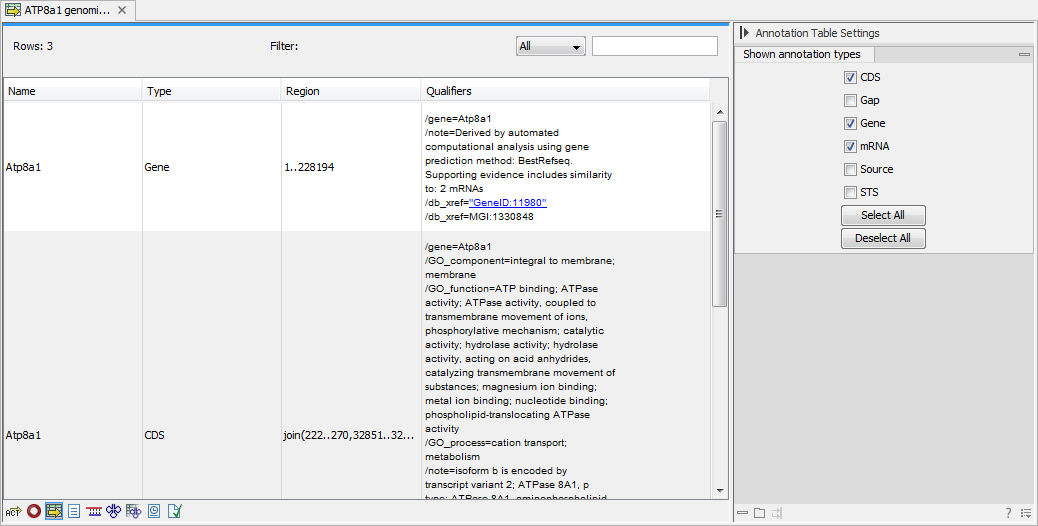
Figure 9.13: A table showing annotations on the sequence.
In the Side Panel you can show or hide individual annotation types in the table. E.g. if you only wish to see "gene" annotations, de-select the other annotation types so that only "gene" is selected.
Each row in the table is an annotation which is represented with the following information:
- Name.
- Type.
- Region.
- Qualifiers.
This information corresponds to the information in the dialog when you edit and add annotations (see Adding annotations).
You can benefit from this table in several ways:
- It provides an intelligible overview of all the annotations on the sequence.
- You can use the filter at the top to search the annotations. Type e.g. "UCP" into the filter and you will find all annotations which have "UCP" in either the name, the type, the region or the qualifiers. Combined with showing or hiding the annotation types in the Side Panel, this makes it easy to find annotations or a subset of annotations.
- You can copy and paste annotations, e.g. from one sequence to another.
- If you wish to edit many annotations consecutively, the double-click editing makes this very fast (see Adding annotations).
