RNA-Seq report
An example of an RNA-seq report that is created if you choose the Create report option is shown in figure 27.16.
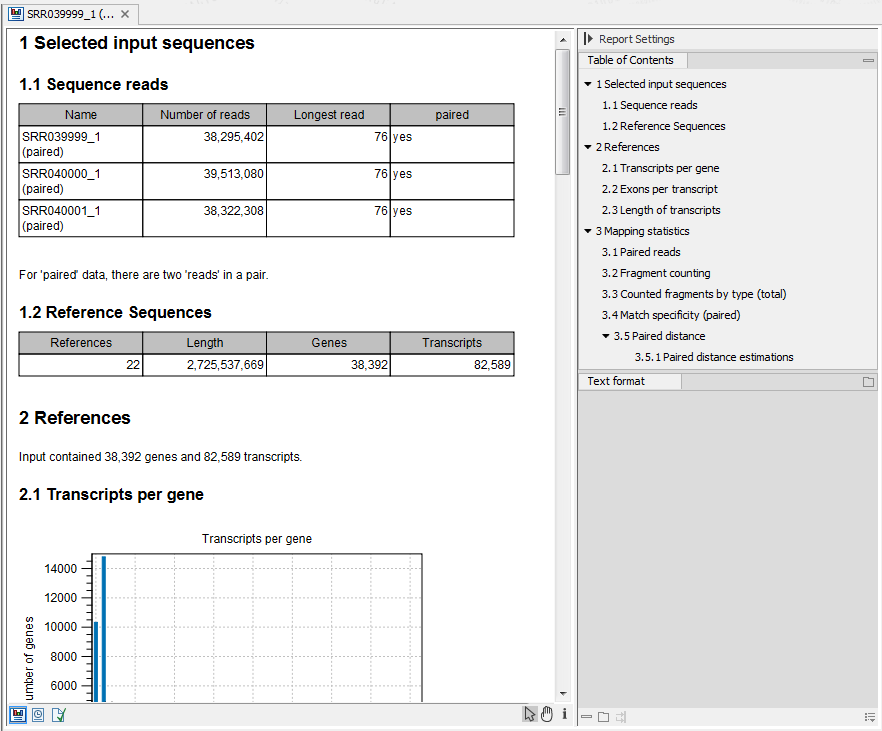
Figure 27.16: Report of an RNA-Seq run.
The report contains the following information:
- Sequence reads. Information about the number of reads.
- Reference sequences. Information about the reference sequences used and their lengths.
- Reference. Information about the total number of genes and transcripts (for eukaryotes only) found in the reference.
- Transcripts per gene. A graph showing the number of transcripts per gene. For eukaryotes, this will be equivalent to the number of mRNA annotations per gene annotation.
- Exons per transcript. A graph showing the number of exons per transcript.
- Length of transcripts. A graph showing the distribution of transcript lengths.
- Mapping statistics. Shows statistics on:
- Paired reads. (Only included if paired reads are used). Shows the number of reads mapped in pairs, the number of reads in broken pairs and the number of unmapped reads.
- Fragment counting. Lists the total number of fragments used for calculating expression, divided into uniquely and non-specifically mapped reads (see the point below on match specificity for details).
- Counted fragments by type. Divides the fragments that are counted into different types
- Exon. Reads that map completely within an exon
- Exon-exon reads. Reads that map across an exon junction as specified in figure 27.13.
- Total exon reads. Number of reads that fall entirely within an exon or in an exon-exon junction.
- Intron. Reads that fall partly or entirely within an intron.
- Total gene reads. All reads that map to the gene.
- Intergenic. All reads that map partly or entirely between genes (will only be shown if the Also map to inter-genic regions option is used).
- Match specificity. Shows a graph of the number of match positions for the reads. Most reads will be mapped 0 or 1 time, but there will also be reads matching more than once in the reference. The maximum number of match positions is limited in the Maximum number of hits for a read setting in figure 27.4. Note that the number of reads that are mapped 0 times includes both the number of reads that cannot be mapped at all and the number of reads that matches to more than the Maximum number of hits for a read parameter.
- Paired distance. (Only included if paired reads are used). Shows a graph of the distance between mapped reads in pairs.
