Ingenuity Variant Analysis for Hereditary Diseases
To start the tool, go to:
Ingenuity Variant Analysis | Ingenuity Variant Analysis for Hereditary Diseases
If you are connected to a server, you will first be asked about where you would like to run the analysis. If you are not connected to a server, the first step is to specify the input for the analysis. The Ingenuity Variant Analysis for Hereditary Diseases tool accepts a single variant track (![]() ) as input. Select the proband variant track, i.e., the individual affected by the disease you are studying, as shown in figure 3.1.
) as input. Select the proband variant track, i.e., the individual affected by the disease you are studying, as shown in figure 3.1.
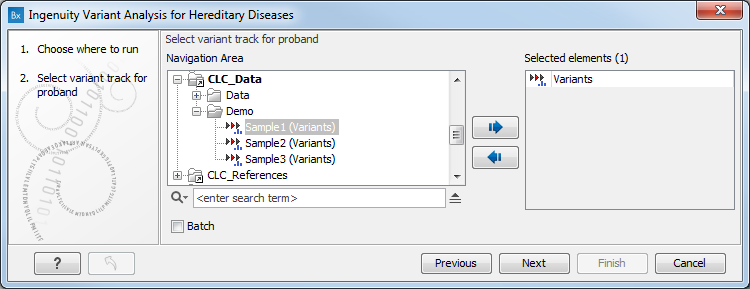
Figure 3.1: Select first the proband variant track.
You can then set the analysis parameters (figure 3.2).
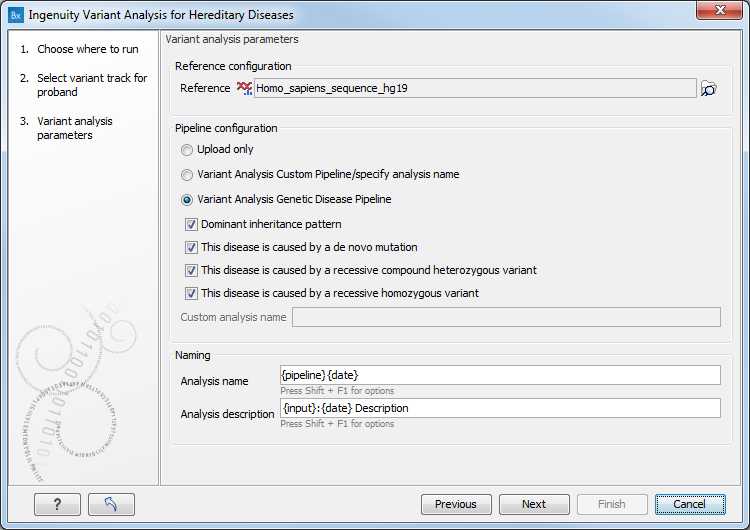
Figure 3.2: Specify the analysis parameters.
- Reference: Select the human reference sequence that is found
under CLC_References in the Navigation Area. Complete human genomes (e.g., hg19 (GRCh37) and hg38 (GRCh38)) and subsets of these (e.g. individual
chromosomes) can be used as references.
- Upload only: useful if you just want to upload samples and do not wish to carry out an analysis. Note: in this case, no results will be downloaded.
- Variant Analysis Custom Pipeline: useful if you have already carried out an Ingenuity Variant Analysis where you had set up a desired filtering cascade and want to re-use the same filtering cascade for a new analysis. If this option is selected, the name of the custom analysis must be specified in the Custom analysis name field. The name you enter in the Custom analysis name field must match the "Name" field of an existing analysis in Ingenuity Variant Analysis exactly as it appears in the Ingenuity Variant Analysis web interface. Note: please provide unique names to all your custom analyses in Ingenuity Variant Analysis: you might not be able to reuse a filtering cascade if its name was already used previously, even in cases where that previous analysis was already deleted.
- Variant Analysis Genetic Disease Pipeline: to be used if you are studying genetic disease. Learn more about the four genetic disease pipelines in Genetic disease pipelines.
- Dominant inheritance pattern
- This disease is caused by a de novo mutation.
- This disease is caused by a recessive compound heterozygous variant.
- This disease is caused by a recessive homozygous variant.
- Analysis name: The name of the analysis. You can enter a name of your own choice by typing in the name, or by using the options that appear when you press Shift + F1. The available shorthand notations are: {input} will be substituted with the name of the input experiment, {date} is substituted with a date stamp, and {pipeline} is substituted by the Analysis pipeline name. The analysis name is the name that is shown on the Ingenuity Variant Analysis page when you choose "My Analyses". The same analysis name can furthermore be used in a Variant Analysis Custom Pipeline, if you specify it in the Custom analysis name field (see above).
- Analysis description: This will be the description of the analysis in Ingenuity Variant Analysis once created. There are a few shorthand notations available: {input} will be substituted with the name of the input experiment, {date} is substituted with a date stamp.
The "Family information" dialog allows you to specify family data for the analysis (figure 3.3).
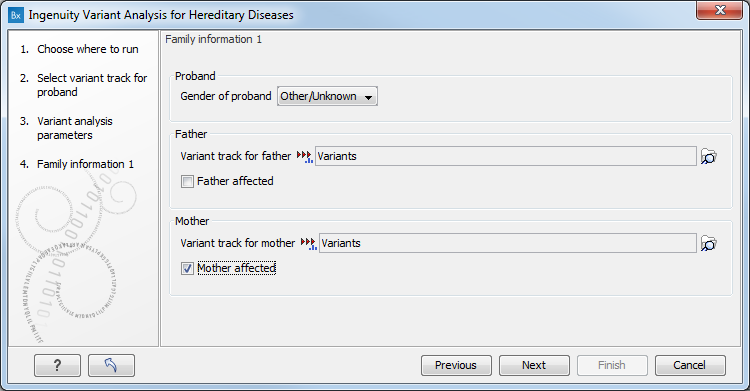
Figure 3.3: Specify family data for your analysis. Data for at least one parent must be specified at this step
- Gender of proband: Select the gender of the individual affected by the disease.
- Variant track for father/mother: Select the variant track for the father or mother, as appropriate. The variant track for at least one parent must be specified.
- Father/mother affected: Once you have selected a variant track for a parent, the option to set the disease status of that parent will be enabled. Check this box if the given parent is affected by the same disease as the proband. Uncheck this box if the given parent is not affected.
A second "Family information" dialog gives you the possibility to specify further family data for the analysis (figure 3.4). Similarly to the previous step, you can specify for each sibling a variant track, disease status and gender. However, check the pipeline of interest in Genetic disease pipelines, as adding non-supported information for a specific pipeline might alter the results.
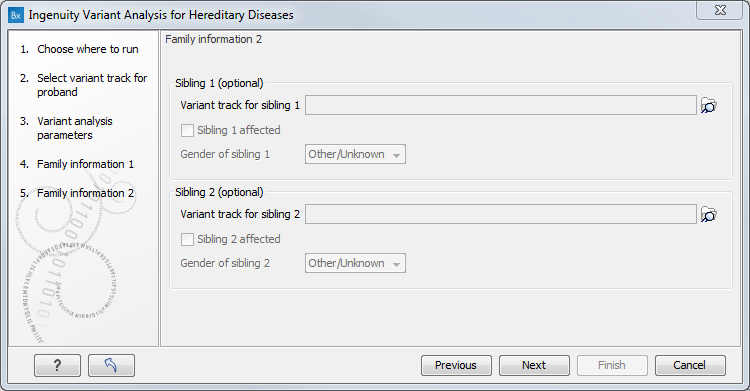
Figure 3.4: At this step, you have the option to specify variant tracks for siblings of the proband.
In the next dialog you must specify your Ingenuity username (email address) and password (figure 3.5). If you do not have an Ingenuity username or password, you must first create an Ingenuity account (see Introduction to the Ingenuity Variant Analysis plugin to learn how).
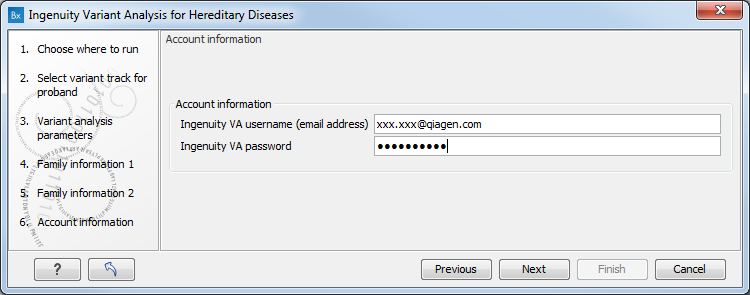
Figure 3.5: Specify the account information: your Ingenuity username (email address) and password are required at this step.
In the "Result handling" wizard step (figure 3.6) you can set the output options.
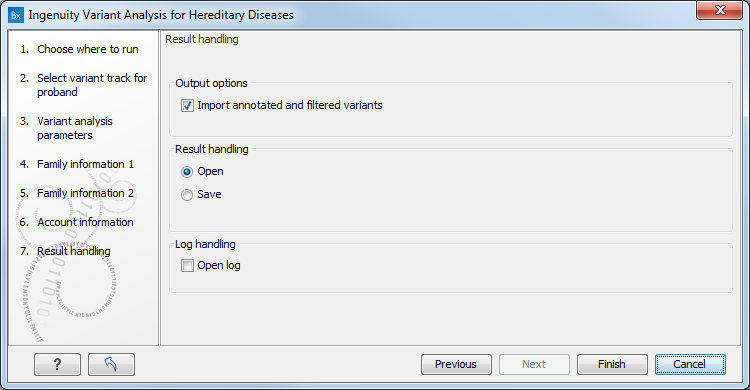
Figure 3.6: The result handling step.
If the Import annotated and filtered variants option is checked, the tool will produce a variant track as output. If it is unchecked, the analysis will be created, and can be accessed inside the Ingenuity Variant Analysis web interface, but the results will not be imported into the workbench. Note: it is not possible to import results if you have selected the Upload only pipeline earlier in the wizard.
If you choose to open the results the two generated outputs will be opened in the View Area without being saved. In this case you will have to manually save the outputs if you would like to keep them. If you choose to save the outputs, click on the button labeled Next to specify where to save the results and click on the button labeled Finish to start the Ingenuity Variant Analysis. Your results will not be opened automatically but will be saved at the destination you have specified.
The outputs and how to manually adjust filter settings are described in Analysis using the plugin and the IVA web interface.
Subsections
