Find Resistance
To perform resistance typing, go to:
Toolbox | Microbial Genomics Module (![]() ) | Typing and Epidemiology (beta) (
) | Typing and Epidemiology (beta) (![]() ) | Find Resistance (
) | Find Resistance (![]() )
)
- Select the input genome or contigs (figure 11.4) and click Next.
Figure 11.4: Pre-assembled and complete- or partial genomes simple contig sequences may be used as input for resistance typing. - Specify the settings for the tool (figure 11.5).
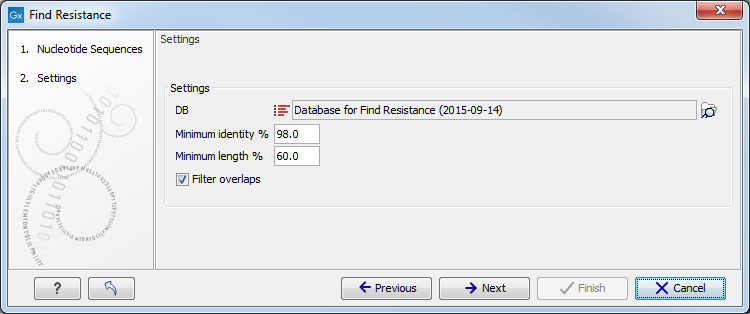
Figure 11.5: Select database and settings for resistance typing.The settings that can be set are
- DB Select the downloaded database for Find Resistance (see section 11.3).
- Minimum Identity % is the threshold for the minimum percentage of nucleotides that are identical between the best matching resistance gene in the database and the corresponding sequence in the genome.
- Minimum Length % reflect the percentage of the total resistance gene length that a sequence must overlap a resistance gene to count as a hit for that gene. Here represented as a percentage of the total resistance gene length.
- Filter overlaps: will perform extra filtering of results that agree on contig, source and predicted phenotype where one is contained by the other.
- Save your data and click on Finish.
The output of the find Resistance tool is a table listing all the possible resistance genes and predicted phenotypes found in the input genome or contigs, as well as additional information such as degrees of similarity between the gene found in the genome and the reference (% identity and query /HSB values), the location where the gene was found (contig name, and position in the contig) as well as a link to NCBI. To add the obtained resistance types to your Result Metadata Table, see the section Add to Result Metadata Table.
