Primer Parameters
In this preference group a number of criteria can be set, which the selected primers must meet. All the criteria concern single primers, as primer pairs are not generated until the Calculate button is pressed. Parameters regarding primer and probe sets are described in detail for each reaction mode (see Standard PCR, Nested PCR, TaqMan or Sequencing primers).- Length. Determines the length interval within which primers can be designed by setting a maximum and a minimum length. The upper and lower lengths allowed by the program are 50 and 10 nucleotides respectively.
- Melting temperature. Determines the temperature interval within which primers must lie. When the Nested PCR or TaqMan reaction type is chosen, the first pair of melting temperature interval settings relate to the outer primer pair i.e. not the probe. Melting temperatures are calculated by a nearest-neighbor model which considers stacking interactions between neighboring bases in the primer-template complex. The model uses state-of-the-art thermodynamic parameters [SantaLucia, 1998] and considers the important contribution from the dangling ends that are present when a short primer anneals to a template sequence [Bommarito et al., 2000]. A number of parameters can be adjusted concerning the reaction mixture and which influence melting temperatures (see below). Melting temperatures are corrected for the presence of monovalent cations using the model of [SantaLucia, 1998] and temperatures are further corrected for the presence of magnesium, deoxynucleotide triphosphates (dNTP) and dimethyl sulfoxide (DMSO) using the model of [von Ahsen et al., 2001].
- Inner melting temperature. This option is only activated when the Nested PCR or TaqMan mode is selected. In Nested PCR mode, it determines the allowed melting temperature interval for the inner/nested pair of primers, and in TaqMan mode it determines the allowed temperature interval for the TaqMan probe.
- Advanced parameters. A number of less commonly used options
- Buffer properties. A number of parameters concerning the reaction mixture which influence melting temperatures.
- Primer concentration. Specifies the concentration of primers and probes in units of nanomoles (
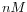 )
)
- Salt concentration. Specifies the concentration of monovalent cations (
![$ [NA^+]$](img10.gif) ,
, ![$ [K^+]$](img11.gif) and equivalents) in units of millimoles (
and equivalents) in units of millimoles (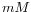 )
)
- Magnesium concentration. Specifies the concentration of magnesium cations (
![$ [Mg^{++}]$](img13.gif) ) in units of millimoles (
) in units of millimoles (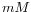 )
)
- dNTP concentration. Specifies the combined concentration of all deoxynucleotide triphosphates in units of millimoles (
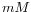 )
)
- DMSO concentration. Specifies the concentration of dimethyl sulfoxide in units of volume percent (
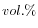 )
)
- Primer concentration. Specifies the concentration of primers and probes in units of nanomoles (
- GC content. Determines the interval of CG content (% C and G nucleotides in the primer) within which primers must lie by setting a maximum and a minimum GC content.
- Self annealing. Determines the maximum self annealing value of all primers and probes. This determines the amount of base-pairing allowed between two copies of the same molecule. The self annealing score is measured in number of hydrogen bonds between two copies of primer molecules, with A-T base pairs contributing 2 hydrogen bonds and G-C base pairs contributing 3 hydrogen bonds.
- Self end annealing. Determines the maximum self end annealing value of all primers and probes. This determines
the number of consecutive base pairs allowed between the 3' end of one primer and another copy of that primer. This score is calculated in number
of hydrogen bonds (the example below has a score of 4 - derived from 2 A-T base pairs each with 2 hydrogen bonds).
AATTCCCTACAATCCCCAAA || AAACCCCTAACATCCCTTAA. - Secondary structure. Determines the maximum score of the optimal secondary DNA structure found for a primer or probe. Secondary structures are scored by the number of hydrogen bonds in the structure, and 2 extra hydrogen bonds are added for each stacking base-pair in the structure.
- Buffer properties. A number of parameters concerning the reaction mixture which influence melting temperatures.
- 3' end G/C restrictions. When this checkbox is selected it is possible to specify
restrictions concerning the number of G and C molecules in the 3'
end of primers and probes. A low G/C content of the primer/probe 3'
end increases the specificity of the reaction. A high G/C content
facilitates a tight binding of the oligo to the template but also
increases the possibility of mispriming. Unfolding the preference
groups yields the following options:
- End length. The number of consecutive terminal nucleotides for which to consider the C/G content
- Max no. of G/C. The maximum number of G and C nucleotides allowed within the specified length interval
- Min no. of G/C. The minimum number of G and C nucleotides required within the specified length interval
- 5' end G/C restrictions. When this checkbox is selected it is possible to specify restrictions concerning the number of G and C molecules in the 5' end of primers and probes. A high G/C content facilitates a tight binding of the oligo to the template but also increases the possibility of mis-priming. Unfolding the preference groups yields the same options as described above for the 3' end.
- Mode. Specifies the reaction type for which primers are
designed:
- Standard PCR. Used when the objective is to design primers, or primer pairs, for PCR amplification of a single DNA fragment.
- Nested PCR. Used when the objective is to design two primer pairs for nested PCR amplification of a single DNA fragment.
- Sequencing. Used when the objective is to design primers for DNA sequencing.
- TaqMan. Used when the objective is to design a primer pair and a probe for TaqMan quantitative PCR.
Each mode is described further in Standard PCR, Nested PCR, TaqMan and Sequencing primers.
- Calculate. Pushing this button will activate the algorithm for designing primers
