Signal Peptide Prediction (SignalP 4.1)
The tool uses SignalP 4.1 [Nielsen et al., 1997,Bendtsen et al., 2004] which is located at https://services.healthtech.dtu.dk/service.php?SignalP-4.1. An active internet connection is required to query SignalP.
To run the signal peptide prediction, go to:
Toolbox | Classical Sequence Analysis (![]() ) | Protein Analysis (
) | Protein Analysis (![]() ) | Signal
Peptide Prediction (SignalP 4.1) (
) | Signal
Peptide Prediction (SignalP 4.1) (![]() )
)
If a sequence was selected before choosing the Toolbox action, this sequence is now listed in the Selected Elements window of the dialog. Use the arrows to add or remove sequences or sequence lists from the selected elements. The SignalP service is limited to 2,000 sequences and 200,000 amino acids for one submission. Each sequence may be no longer than 6,000 amino acids.
Click Next to set parameters for the SignalP analysis.
You should select which organism group the input sequences belong to. the default is eukaryote (see figure 2.2).
- Eukaryote (default)
- Gram-negative bacteria
- Gram-positive bacteria
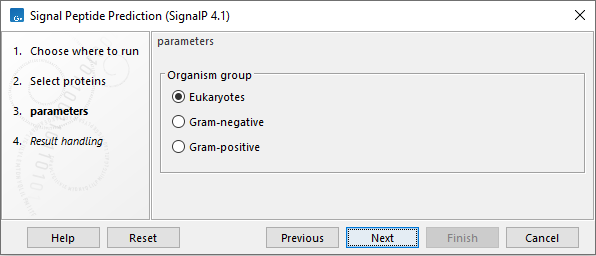
Figure 2.2: Setting the parameters for signal peptide prediction.
You can perform the analysis on several protein sequences at a time. This will add annotations to all the sequences and open a view for each sequence if a signal peptide is found. If no signal peptide is found in the sequence, a dialog box will be shown.
The predictions obtained can either be shown as annotations on the sequence, listed in a table or be shown as the detailed and full text output from the SignalP method. This can be used to interpret borderline predictions:
- Add annotations to sequence
- Create table
- Text
In order to predict potential signal peptides of proteins, the D-score from the SignalP output is used for discrimination of signal peptide versus non-signal peptide (see section 2.1.1). This score has been shown to be the most accurate [Klee and Ellis, 2005] in an evaluation study of signal peptide predictors.
After running the prediction as described above, the protein sequence will show predicted signal peptide as annotations on the original sequence (see figure 2.3). Make sure the Side Panel settings of the sequence is so that 'Show annotations' is checked in the 'Annotation layout' palette, and that the annotation type 'Signal peptide' is checked in the 'Annotation types' palette.
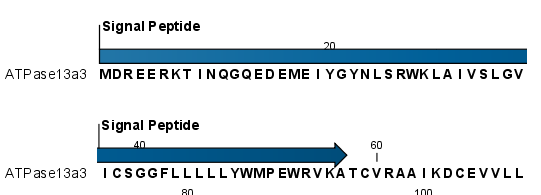
Figure 2.3: N-terminal signal peptide shown as annotation on the sequence.
Additional notes can be added through the Edit
annotation (![]() ) right-click mouse menu. Undesired annotations can be removed through the Delete
Annotation (
) right-click mouse menu. Undesired annotations can be removed through the Delete
Annotation (![]() ) right-click mouse menu.
) right-click mouse menu.
Subsections
