Transmembrane Helix Prediction (DeepTMHMM)
Many proteins are integral membrane proteins. Most membrane proteins have hydrophobic regions which span the hydrophobic core of the membrane bi-layer and hydrophilic regions located on the outside or the inside of the membrane. Many receptor proteins have several transmembrane helices spanning the cellular membrane.
The prediction of transmembrane helices using Transmembrane Helix Prediction (DeepTMHMM), uses the DeepTMHMM service located at https://dtu.biolib.com/DeepTMHMM. Additional information on DeepTMHMM can be found at https://dtu.biolib.com/DeepTMHMM and in the original research paper [Hallgren et al., 2022].
To run a Transmembrane Helix Prediction (DeepTMHMM) analysis from the Workbench, go to:
Tools | Classical Sequence Analysis (![]() ) | Protein Analysis (
) | Protein Analysis (![]() ) |
Transmembrane Helix Prediction (DeepTMHMM) (
) |
Transmembrane Helix Prediction (DeepTMHMM) (![]() )
)
In the first wizard step, you select the peptide sequences to be analyzed.
In the Result handling wizard step, you specify the form the results should be returned in. The options are:
- Add annotations Annotate the input sequences with the results. See figure 3.1 for an example. Note: relevant options must be checked in the Side Panel settings, in the 'Annotation layout' palette, for these annotations to be visible (further details below).
- Create text Get the text output from the DeepTMHMM service. A single text file is created, containing the results returned.
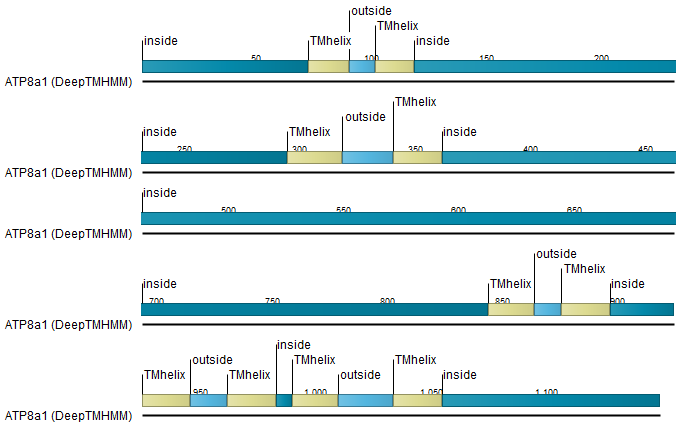
Figure 3.1: Transmembrane segments shown as annotation on the sequence, including information about the the topology.
Hover the mouse cursor over an annotation to reveal a tooltip with details about it. If the annotation was added using DeepTMHMM, this is noted. The annotation types added using DeepTMHMM are easily discovered by opening the Annotation Table view of the sequence, selecting all the available annotation types in the Side Panel. Information in the Qualifiers column includes "DeepTMHMM" for annotations added using this service. Working with sequence annotations is described in detail at https://resources.qiagenbioinformatics.com/manuals/clcmainworkbench/current/index.php?manual=Working_with_annotations.html.
